November 2, 2015 report
Using a DNA scaffold to place molecules with Bohr's radius resolution
(Phys.org)—A new study demonstrates that researchers can control the distance between two molecules such that they can adjust the step size to as small as Bohr's radius. This proof-of-concept study using DNA origami techniques shows how molecular positioning can be fine-tuned at the atomic level at room temperature in solution. This work has applications for molecular architecture as well as templated chemical reactions. This study appears in Nature Nanotechnology.
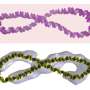
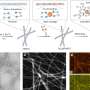
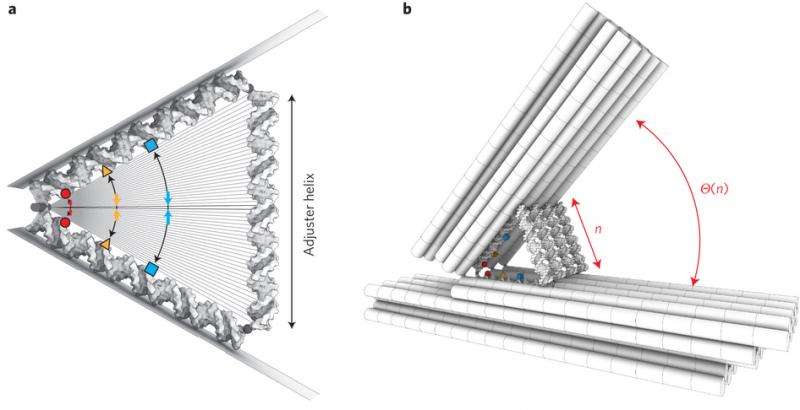
The TEM images in Jonas J. Funke and Hendrik Dietz's report look like a series of simple machines that a student would learn in school, but these simple machines are made of DNA. But, similar to simple machines, as the angle between intersecting pieces increases, the distance between distal endpoints on the intersecting pieces increases. Funke and Dietz controlled the distance of the distal endpoints suing an adjuster helix, a DNA helix whose length is increased by adding base pairs.
The intersecting pieces are also DNA helices meaning that as the angle converges, the distance between one helix and the other decreases. The base pairs on each helix are a certain distance apart from the base pairs on the other helix. As this work demonstrates, that distance is adjustable.
Funke and Dietz demonstrated that the angle changes with increasing length of adjuster helix by making helices of lengths from ten base pairs to fifty base pairs. TEM studies showed a smoothly increasing angle as length of the adjuster helix increased. The DNA arms and adjuster helices provide the scaffold to control distance between two interacting molecules placed on the arms.
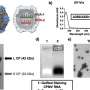
Funke and Dietz used FRET studies to gain a better understanding of the distance and interaction between two molecules on this DNA scaffold. In FRET a donor chromophore transfers energy to an acceptor chromophore. The efficiency of this transfer is related to distance between the chromophores. In this study, chromophores were placed at positions five, fifteen, and twenty-five along the DNA arms. Position five is closest to the vertex angle and twenty-five is farthest away from the vertex. They found a relationship between emission intensities and the length of the adjuster helix. Additionally, electrophoretic studies showed that functionalizing the DNA scaffold with chromophores did not change the properties of the scaffold.
As chromophore distances decreased from 9.0 nm to 3.5 nm, they observed the expected donor/acceptor interactions. When distances decreased from 3.5 nm to 1.5 nm, they observed fluorescence quenching. Their data suggested that they could discern distances as small as 0.04 nm. This was confirmed with an improved study of fluorescence quenching, showing it is possible to discern distances that differ by 0.04 nm, or less than Bohr's radius.
Finally, to understand how thermal fluctuations at room temperature affect molecular distances, Funke and Dietz looked at chemical crosslinking reactions of thiol groups. The thiol groups were placed fifteen base pairs away from the vertex angle and were reacted with five different homo-bifunctional bismaleimide linker molecules with known distances and thermal fluctuations. This allowed for two possible reactions, the crosslinked reaction and the non-crosslinked reaction.
By graphing crosslinking yield as a function of distance, they found that as long as the bismaleimide was long enough to span the distance, then it would produce the crosslinked product. If not, the yield went to zero. Experimental results showed that the yield drop off was gradual and at a larger distance than the contour length of the bismaleimide, due to fluctuations at room temperature and in solution. For example, BMOE, one of the bismaleimide compounds, has a contour length of 1.05 nm, but yield dropped off at 3.5 nm. Using a quantitative model for the reaction, Funke and Dietz were able to calculate the fluctuations in the distance coordinate to within 0.5 nm.
This proof-of-concept study demonstrates the possibility of using a DNA scaffold to control molecular distance. When we asked about the implications of his research, Dr. Funke said, "Arranging matter with ever more precision is a key goal for science and technology. Our study shows, that scaffolded DNA origami enables the rational positioning of two molecules with atomic resolution and therefore opens up new opportunities to study and manipulate molecular interactions.
More information: Jonas J. Funke et al. Placing molecules with Bohr radius resolution using DNA origami, Nature Nanotechnology (2015). DOI: 10.1038/nnano.2015.240
Abstract
Molecular self-assembly with nucleic acids can be used to fabricate discrete objects with defined sizes and arbitrary shapes. It relies on building blocks that are commensurate to those of biological macromolecular machines and should therefore be capable of delivering the atomic-scale placement accuracy known today only from natural and designed proteins. However, research in the field has predominantly focused on producing increasingly large and complex, but more coarsely defined, objects and placing them in an orderly manner on solid substrates. So far, few objects afford a design accuracy better than 5 nm, and the subnanometre scale has been reached only within the unit cells of designed DNA crystals. Here, we report a molecular positioning device made from a hinged DNA origami object in which the angle between the two structural units can be controlled with adjuster helices. To test the positioning capabilities of the device, we used photophysical and crosslinking assays that report the coordinate of interest directly with atomic resolution. Using this combination of placement and analysis, we rationally adjusted the average distance between fluorescent molecules and reactive groups from 1.5 to 9 nm in 123 discrete displacement steps. The smallest displacement step possible was 0.04 nm, which is slightly less than the Bohr radius. The fluctuation amplitudes in the distance coordinate were also small (±0.5 nm), and within a factor of two to three of the amplitudes found in protein structures.
Journal information: Nature Nanotechnology
© 2015 Phys.org