September 15, 2015 report
Sculpting a conjugated polymer using DNA origami
(Phys.org)—New research allows scientists to sculpt polymers into two- and three-dimensional shapes, similar to how polypeptides fold into functional three-dimensional shapes. This ability is particularly advantageous for conjugated polymers, polymers that have a networked pi-electron system, because they are conducting. Immobilizing and shaping conducting polymers is an important step in constructing molecular circuits.
A group of scientists from Aarhus University in Denmark, the Wyss Institute at Harvard, and Max Plank Institute in Germany, have synthesized, characterized, and immobilized a conjugated polymer using DNA origami. Their polymer was able to be shaped and molded into various two- and three-dimensional shapes while maintaining its physical properties. Their work appears in Nature Nanotechnology.
Knudsen, et al. synthesized a conjugated brush polymer, (2,5-dialkoxy) paraphenylene vinylene (APPV) that is functionalized with a nine nucleotide long single stranded DNA (ssDNA) sequences to serve as a linkage to the DNA origami. APPV has hydroxyl groups along its backbone that are attached to phenylene units. These hydroxyl groups are available for functionalization with synthetic ssDNA.
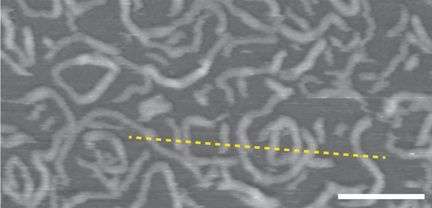
The ssDNA annealed to complementary strands that extend from a DNA platform, thus holding the polymer in place. This technique is known as DNA origami because the complementary strands of DNA extending from the DNA origami can be tailored to any shape or design and should guide the polymer with its complementary ssDNA to take that same shape.
In this experiment, APPV-DNA was characterized with gel permeation chromatography, UV-Vis spectroscopy, fluorescence spectroscopy, XPS, and AFM. Gel permeation chromatography showed that the polymer size ranged from 340 kDa to 3,300 kDa. This and AFM studies indicated the presence of smaller and longer polymer pieces. XPS showed that less than two-thirds of the phenylene units, containing hydroxyl groups, where functionalized with ssDNA. Additionally, AFM studies provided surface potential information, indicating that the APPV-DNA polymer has a higher charge transfer than the silicon oxide substrate but lower than gold or carbon nanotubes.
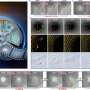
The polymer was then immobilized on DNA origami into various two and three-dimensional shapes, and charge transfer as well as polymer integrity were tested. The first test involved DNA origami in linear, U-shaped, and at 90o angles. Surface potential studies indicated that the immobilized APPV-DNA polymer displayed similar charge transfer abilities in all conformations. The polymer's flexibility was verified by subjecting it to DNA origami shapes that would strain the structure: wave, staircase, and circular.
Finally, the APPV-DNA polymer was formed into a three-dimensional cylindrical structure made from stacked rings of double helices. The stacked rings are held together using staple strands. TEM studies confirmed the shape of the cylinder, but the polymer does not provide adequate contrast for full characterization using TEM. AFM or any other scanning microscopy techniques will not work for this kind of structure either. The interaction between the tip and the molecule could damage the "soft" three-dimensional structure of the polymer.
In order to obtain a three-dimensional rendering of the APPV-DNA cylinder, Knudsen, et al. used DNA-PAINT. Using the excess nine-nucleotide ssDNA that did not bind to the DNA origami structure, Knudsen, et al. made complementary strands with a fluorescent label. They then used DNA-PAINT to visualize the strand pattern and render a three-dimensional image.
This research demonstrates the ability to control the two and three-dimensional conformation of a conjugated polymer, which has promising implications for molecular circuit design.
More information: "Routing of individual polymers in designed patterns" Nature Nanotechnology, DOI: 10.1038/NNANO.2015.190
Abstract
Synthetic polymers are ubiquitous in the modern world, but our ability to exert control over the molecular conformation of individual polymers is very limited. In particular, although the programmable self-assembly of oligonucleotides and proteins into artificial nanostructures has been demonstrated, we currently lack the tools to handle other types of synthetic polymers individually and thus the ability to utilize and study their single-molecule properties. Here we show that synthetic polymer wires containing short oligonucleotides that extend from each repeat can be made to assemble into arbitrary routings. The wires, which can be more than 200 nm in length, are soft and bendable, and the DNA strands allow individual polymers to self-assemble into predesigned routings on both two- and three-dimensional DNA origami templates. The polymers are conjugated and potentially conducting, and could therefore be used to create molecular-scale electronic or optical wires in arbitrary geometries.
Journal information: Nature Nanotechnology
© 2015 Phys.org