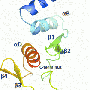Viral protein in their sights: Advanced imaging reveals key structure of Ebola and other RNA viruses
Viruses need us. In order to multiply, viruses have to invade a host cell and copy their genetic information. To do so, viruses encode their own replication machinery or components that subvert the host replication machinery to their advantage.
Ebola virus and rabies virus, two of the most lethal pathogens known to humans, belong to an order of RNA viruses that share a common strategy for copying their genomes inside their hosts. Other relatives include Marburg virus, measles, mumps, respiratory syncytial virus and vesicular stomatitis virus (VSV). Scientists study VSV, which causes acute disease in livestock but typically does not lead to illness in people, as a model for viruses that are harmful to humans.
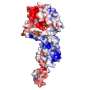
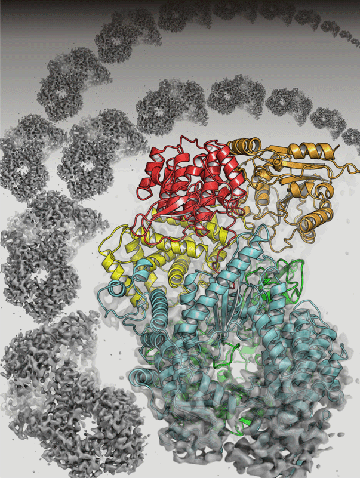
Now a team from Harvard Medical School, using electron cryomicroscopy (imaging frozen specimens to reduce damage from electron radiation), has for the first time revealed the structure of a VSV protein at the atomic level. Called polymerase protein L, it is required for viral replication in this group of RNA viruses. The findings are published in Cell.
"We now have a better understanding of how RNA synthesis works for these viruses," said Sean Whelan, HMS professor of microbiology and immunobiology and senior author of the paper. "I think if you were trying to develop a viral-specific target to block the replication of one of these viruses, having the structure of the polymerase protein would help."
Scientists already know how these RNA viruses infect cells. They start by delivering a large protein RNA complex, which is viral RNA enclosed in a protein coat. The protein that copies viral RNA is polymerase protein L, which conducts all the enzymatic activities needed to synthesize RNA and then add a cap structure to its end to ensure it doesn't get destroyed by the cell—and to ensure that it can be translated into protein.
While researchers have known the atomic structures of the protein that coats the viral RNA, there are no data on protein L's atomic structure.
Antiviral drugs that target polymerase molecules are based in part on knowing their structure. That approach has been successful against HIV and herpes and hepatitis C viruses. But for the class of viruses known as nonsegmented negative-strand RNA viruses, finding the structure of polymerase protein L has been challenging.
The "L" stands for large. Larger proteins are often difficult to produce and to purify, Whelan said. Protein L is also flexible, with many functional fragments that are hard to isolate. The viruses evolved to make only small quantities of this protein.
Five years ago, using a lower-resolution form of electron microscopy in which the protein is visualized in the presence of negative stain, Whelan's team was able to detect at low resolution a structure that looked like a doughnut with three globular domains. Those earlier studies were informative, but the approach could not provide the atomic level of resolution the team ultimately needed.
Advances in electron cryomicroscopy encouraged them to try again. A team from Whelan's lab, working with a group led by Stephen Harrison, Giovanni Armenise - Harvard Professor of Basic Biomedical Science at HMS and a Howard Hughes Medical Institute (HHMI) investigator, was able to collect data from their viral samples that gave them much greater resolution. They also were able to align the images they collected into a three-dimensional model of polymerase protein L.
Into the density map obtained from these studies, members of the team built an atomic model of the polypeptide chain of VSV L protein. Solving this puzzle was a significant challenge and also involved the team of Nikolaus Grigorieff at HHMI's Janelia campus.
The result? An atomic level model of polymerase protein L's structure for VSV, which will form the basis for understanding the L protein of the other viruses in the order.
"The Ebola protein will look the same, the rabies protein will look the same, the other L proteins will look the same," Whelan said. "There will be some subtle differences reflecting the precise nature of amino acids, but we know that they're functionally and structurally the same."
Knowing the structure means scientists can explore how RNA synthesis is working in these viruses.
"It begins to suggest ways that we can perhaps pull apart other proteins that have not been so easy to express, such as the L protein in Ebola," Whelan said. "It doesn't mean we're going to have inhibitors immediately, but this is an important step, I think, towards that longer-term goal."
Journal information: Cell
Provided by Harvard Medical School