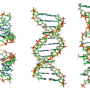Epigenetic 'switch' regulates RNA-protein interactions
Chemical changes - also known as epigenetic modifications - to messenger RNA (mRNA) are thought to play an important role in gene expression, and have recently been found to affect biological processes such as circadian clock management and obesity. But the specific mechanisms involved have been poorly understood.
A new study by scientists from the University of Chicago now shows that epigenetic modifications to mRNA act as a structural "switch" that - by physically opening space in a folded-up strand of mRNA -allows RNA-binding proteins to recognize and read mRNA regions that would otherwise be inaccessible. The findings, reported in Nature on Feb. 25, provide a new understanding of this emerging field of study.
"RNA epigenetic modifications affect practically all RNA-protein interactions," said senior study author Tao Pan, PhD, professor of biochemistry and molecular biology at the University of Chicago. "This 'switch' mechanism is expected to work as a master regulator of wide-ranging biological activities through influencing RNA-protein interactions."
The expression of genes is regulated by several different mechanisms. One such mechanism, epigenetic changes to DNA, has been well studied for how it contributes to the activation or deactivation of genes. Epigenetic changes to RNA have also been identified, but only recently has this phenomenon - broadly known as the epitranscriptome - been scrutinized for its role in gene expression.
The most abundant epigenetic change to RNA is the addition of a methyl group to certain adenosine nucleotides, known as N6-methyl-adenosine (m6A) modification. This is thought to represent a "code" that affects how RNA-binding proteins read and interpret the genetic instructions conveyed by RNA.
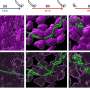
Pan and his colleagues identified specific m6A modifications on an mRNA that increased its interaction with a common RNA-binding protein known as HNRNPC. This protein recognizes and binds to only a specific sequence of nucleotides - five uridines in a row. However, m6A modification only alters adenosine nucleotides. Direct binding to the m6A site by HNRNPC could not explain differences in mRNA-protein interaction.
Unlike the double helix structure of DNA, single-stranded mRNA folds itself into varying and complex structures. The researchers first found that a specific m6A modification they identified was at a location where the RNA strand folded up on itself into a hairpin loop. Here, directly opposite to the m6A site, was a sequence of five uridines in a row - the target binding site for HNRNPC.
The team discovered that the presence of m6A made the target binding site more accessible by physically opening a space for the protein to recognize the site. This increased the affinity of HNRNPC for that RNA. Without m6A, the binding site was hidden within the hairpin and inaccessible.
"RNA-binding proteins such as HNRNPC can have hundreds of thousands of potential binding sites in a cell," Pan said. "Methylation in the RNA structure allows sites that would otherwise be buried to better compete for binding proteins. We call this the m6A switch."
The team verified their findings through a model and an analytical method that allowed them to determine that these uridine sites were reliant on m6A to bind to HNRNPC. They also applied a method to search for these sites throughout the cell, and found almost 40,000 such m6A sites that affected HNRNPC binding, indicating that this phenomenon was widespread and likely could be applied for other protein interactions.
The researchers found that when m6A was removed from these sites, the abundance of the mRNA was lowered, and that there were changes to alternative splicing patterns. "The implication is, of course, that epigenetic changes to the mRNA ultimately change the protein that's created from it, which could be connected to health or disease," Pan said.
The team is now further analyzing the structural and functional consequences of the m6A switch, as well as mutations that affect its function. "We're still learning much about how genes are regulated," Pan adds. "This mechanism represents another layer of epigenetic regulation to gene expression, analogous to DNA methylation and histone modification."
More information: Nature DOI: 10.1038/nature14234
Journal information: Nature
Provided by University of Chicago Medical Center