Harnessing data from Nature's great evolutionary experiment
There are 3 billion letters in the human genome, and scientists have endlessly debated how many of them serve a functional purpose. There are those letters that encode genes, our hereditary information, and those that provide instructions about how cells can use the genes. But those sequences are written with a comparative few of the vast number of DNA letters. Scientists have long debated how much of, or even if, the rest of our genome does anything, some going so far as to designate the part not devoted to encoding proteins as "junk DNA."
In work published today in Nature Genetics, researchers at Cold Spring Harbor Laboratory (CSHL) have developed a new computational method to identify which letters in the human genome are functionally important. Their computer program, called fitCons, harnesses the power of evolution, comparing changes in DNA letters across not just related species, but also between multiple individuals in a single species. The results provide a surprising picture of just how little of our genome has been "conserved" by Nature not only across species over eons of time, but also over the more recent time period during which humans differentiated from one another.
"In model organisms, like yeast or flies, scientists often generate mutations to determine which letters in a DNA sequence are needed for a particular gene to function," explains CSHL Professor Adam Siepel. "We can't do that with humans. But when you think about it, Nature has been doing a similar experiment on a very large scale as species evolve. Mutations occur across the genome at random, but important letters are retained by natural selection, while the rest are free to change with no adverse consequence to the organism."
It was this idea that became the basis of their analysis, but it alone wasn't enough. "Massive research consortia, like the ENCODE Project, have provided the scientific community with a trove of information about genomic function over the last few years," says Siepel. "Other groups have sequenced large numbers of humans and nonhuman primates. For the first time, these big data sets give us both a broad and exceptionally detailed picture of both biochemical activity along the genome and how DNA sequences have changed over time."
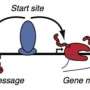
Siepel's team began by sorting ENCODE consortium data based on combinations of biochemical markers that indicate the type of activity at each position. "We didn't just use sequence patterns. ENCODE provided us with information about where along the full genome DNA is read and how it is modified with biochemical tags," says Brad Gulko, a Ph.D. student in Computer Science at Cornell University and lead author on the new paper. The combinations of these tags revealed several hundred different classes of sites within the genome each having a potentially different role in genomic activity.
The researchers then turned to their previously developed computational method, called INSIGHT, to analyze how much the sequences in these classes had varied over both short and long periods of evolutionary time. "Usually, this, kind of analysis is done comparing different species - like humans, dogs, and mice - which means researchers are looking at changes that occurred over relatively long time periods," explains Siepel. But the INSIGHT model considers the changes among dozens of human individuals and close relatives, such as the chimpanzee, which provides a picture of evolution over much shorter time frames.
The scientists found that, at most, only about 7% of the letters in the human genome are functionally important. "We were impressed with how low that number is," says Siepel. "Some analyses of the ENCODE data alone have argued that upwards of 80% of the genome is functional, but our evolutionary analysis suggests that isn't the case." He added, "other researchers have estimated that similarly small fractions of the genome have been conserved over long time evolutionary periods, but our analysis indicates that the much larger ENCODE-based estimates can't be explained by gains of new functional sequences on the human lineage. We think most of the sequences designated as 'biochemically active' by ENCODE are probably not evolutionarily important in humans."
According to Siepel, this analysis will allow researchers to isolate functionally important sequences in diseases much more rapidly. Most genome-wide studies implicate massive regions, containing tens of thousands of letters, associated with disease. "Our analysis helps to pinpoint which letters in these sequences are likely to be functional because they are both biochemically active and have been preserved by evolution." says Siepel. "This provides a powerful resource as scientists work to understand the genetic basis of disease."
More information: "A method for calculating probabilities of fitness consequences for point mutations across the human genome" appears online in Nature Genetics on January 19, 2015. The authors are: Brad Gulko, Melissa Hubisz, Ilan Gronau, and Adam Siepel. The paper can be obtained online at: dx.doi.org/10.1038/ng.3196
Journal information: Nature Genetics
Provided by Cold Spring Harbor Laboratory