New sequence of naked mole rat genome facilitates cancer resistance research
Director of Science at The Genome Analysis Centre (TGAC) Federica Di Palma co-authors new genetic study on the naked mole rat's resistance to cancer, identifying key genomic variations that may have contributed to the evolution of this extraordinary species.
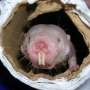
The naked mole rat is an exceptionally long-lived and cancer-resistant rodent native to East Africa. The new study presents a higher-quality assembly of the rodent's genetic structure to previous sequences of the species genome, enabling the research community to benefit from this key data.
The study, led by international scientists from TGAC, University of Liverpool, Broad Institute, Uppsala University and Harvard Medical School, re-analysed the naked mole rat genome using the improved assembly that revealed further candidate genes of potential relevance to adaptive changes in the context of aging and cancer.
With a life span of over thirty years, not only is the naked mole rat (Heterocephalus glaber) the longest-lived rodent, but it is also extremely resistant to neoplasia (tumours), and therefore is an ideal model for research on longevity, cancer and disease resistance.
Senior author from the University of Liverpool's Institute of Integrative Biology, Dr Joao Pedro De Magalhaes, said: "The new study provides a fundamental resource for research on the naked mole rat and its many evolutionary adaptations, including longevity and resistance to diseases, as well as other traits (metabolic regulation, development, pain, and behaviour). We predict that studying a species so long-lived (particularly given its small body size) and with such an astonishing resistance to neoplasia, will help elucidate mechanisms and genes conferring longevity and cancer resistance in mammals that may have human applications."
To help facilitate and encourage further research into this fascinating species, the team of scientists have developed a freely-available online portal, the Naked Mole-Rat Genome Resource (www.naked-mole-rat.org), featuring the new genome sequence and the data results of their analysis.
Federica Di Palma, co-author and Director of Science at TGAC, said: "A high-quality, annotated naked mole rat genome is essential for the research community to develop the sophisticated molecular biology tools necessary to study these amazing animals. By creating a genome resource for the naked mole rat with an advanced genome assembly, we aim to facilitate studies into this fascinating animal and help establish the naked mole rat as the first long-lived model for bioscience research underpinning health."
More information: Michael Keane, Thomas Craig, Jessica Alföldi, Aaron M. Berlin, Jeremy Johnson, Andrei Seluanov, Vera Gorbunova, Federica Di Palma, Kerstin Lindblad-Toh, George M. Church, and João Pedro de Magalhães. "The Naked Mole Rat Genome Resource: facilitating analyses of cancer and longevity-related adaptations." Bioinformatics first published online August 28, 2014 DOI: 10.1093/bioinformatics/btu579
Journal information: Bioinformatics
Provided by The Genome Analysis Centre (TGAC)