Mathematical modelling disproves long-held view of bacterial cell cycle
A key theory of the cell cycle of asymmetric bacteria, which has prevailed for the last ten years, has been disproved by a combined approach using mathematical modelling and genetic experiments.
Modellers Prof. Martin Howard and Dr Seán Murray, from the John Innes Centre on the Norwich Research Park, together with experimental collaborators Prof. Patrick Viollier, Dr Gaël Panis and Coralie Fumeaux at the University of Geneva, had been researching how viruses could hijack the natural cell cycle of Caulobacter crescentus, a bacterium found in water.
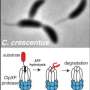
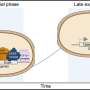
Caulobacter divides asymmetrically – a stationary 'mother' cell divides to produce a motile daughter cell that, unlike the mother, can swim by using a rotating hair-like structure called a flagellum. Unable to reproduce as it is, the daughter cell must first develop into a new mother cell, shedding its flagellum and gaining a stalked appendage that binds it to a surface. This kind of asymmetric life cycle can be seen across nature and is the same type of mechanism used by many multi-cellular organisms like coral.
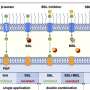
But while developing and testing simple mathematical models of the Caulobacter cell cycle to study its interaction with viruses, they realised that two of four key cell cycle genes thought to be essential for cell viability, were actually not needed at all. Moreover, these two inessential genes turned out to form a regulatory circuit that was conserved in many other asymmetric bacteria, but which was likely absent from Caulobacter's original (and simple) evolutionary ancestors.
"The prevailing view was that without all four of these core cell cycle genes, the cells would not develop properly and would die," said Prof Howard. "Through mathematical modelling, coupled with biological experiments from our colleagues in Geneva, we have shown that prevailing view to be false, as we were able to delete two of the four genes and still obtain viable cells."
The machinery at the heart of asymmetric cell cycle control is thus far simpler than previously believed. This simplicity, and the combined minimal modelling/genetics approach which revealed it, are potentially important for many other cell cycles in more complex organisms. These results have been published 31 December 2013 in PLoS Biology.
Journal information: PLoS Biology
Provided by John Innes Centre