Microbial genomes help propose phylum name

At the phylum level, the number and diversity of unknown microbes still far outnumber those being studied. Metagenomics and single-cell genomics are tools helping researchers learn more about the "biological dark matter" that has not been cultivated and studied in the laboratory.
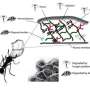
In an article published May 14, 2013 in Nature Communications, a team including DOE Joint Genome Institute researchers and collaborators used both techniques to assemble two nearly-complete genomes belonging to the candidate phylum OP9, which was first discovered in the Obsidian Pool at Yellowstone National Park. The findings have led the team to suggest the phylum be called "Atribacteria."
The OP9 genomes were assembled from single cells collected at a hot spring in California and a metagenomic data set from cellulosic biomass communities being cultivated in Nevadan hot spring sediments. Based on the team's analyses of the microbial genomes' cell structures and central metabolisms, they proposed naming the genomes "Candidatus Caldatribacterium californiense" and "Ca. Caldatribacterium saccharofermentans" respectively. Their findings revealed that OP9 may play a role in breaking down and using (hemi)cellulose.
"This study also illustrates the utility of combining single-cell and metagenomics approaches, even in cases where data sets originate in distinct species or environments," they added.
More information: www.nature.com/ncomms/journal/ … full/ncomms2884.html
Journal information: Nature Communications
Provided by DOE/Joint Genome Institute