How proteins read meta DNA code
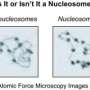
Three-quarters of the DNA in evolved organisms is wrapped around proteins, forming the basic unit of DNA packaging called nucleosomes, like a thread around a spool. The problem lies in understanding how DNA can then be read by such proteins. Now, Arman Fathizadeh, a physicist at Sharif University of Technology in Tehran, Iran, and colleagues have created a model showing how proteins move along DNA, in a paper just published in European Physical Journal E.
The problem is that until now, we did not clearly understand the physical mechanisms of how to "open the book" to read the genetic text contained in DNA. Studying the dynamics of the nucleosome over reasonable time scales by means of molecular dynamics simulations is out of the question, as it would be too complex. Instead, the authors developed a basic computer model of the nucleosome in which DNA is described by a sequence of rigid blocks representing the base pairs. By introducing flexible binding sites of the DNA to the protein core, it provides a more physical representation of the system. It also makes it possible to identify the sliding mechanism of nucleosomes along the DNA.
The idea is that a small defect in the form of a missing or extra base pair enters the DNA section wrapped around a nucleosome. This defect can then diffuse through the wrapped DNA and once it leaves the other end of the wrapped section, the nucleosome moves by the extra or missing length that the defect carried with it. This model supports the idea of a second genetic code, previously suggested in 2006. This would consist of a mechanical code written down within the base pair sequence and multiplexed with the traditional genetic code.
More information: A. Fathizadeh et al. (2013), Rigid body molecular dynamics of DNA inside a nucleosome, European Physical Journal E (2013) 36: 21, DOI 10.1140/epje/i2013-13021-4
Journal information: European Physical Journal E
Provided by Springer