Chasing a common cold virus
(Phys.org)—As the cold and flu season makes its annual visit, a team of researchers, using Argonne's Advanced Photon Source, continue to complete a detailed map of the human adenovirus—one of several viruses responsible for the common cold.
Although human adenovirus is usually not deadly, it is a seasonal nuisance to some and can cause deadly infections in others.
In the early 1990s, Glen Nemerow, professor and principal investigator at the Immunology and Microbial Science division of The Scripps Institute, switched from his previous research on Epstein-Barr to human adenovirus. "It is easy to grow and manipulate, which made (and makes) it an excellent model for studying host-cell interactions," said Nemerow.
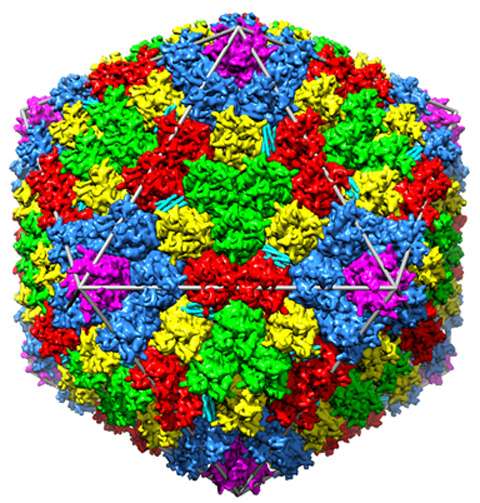
Although the virus had been studied since the early 1950s, most research tended to focus on subunits of the virus, such as major proteins, because the intact virus is dauntingly large. Nemerow decided instead to examine the entire molecular structure of human adenovirus. He wanted to see how proteins self-assemble to form the complete virus. A major goal of the research is to try to improve how the virus could be used for gene delivery—and potentially, cancer therapy. Nemerow teamed up with Vijay Reddy, an associate professor at Scripps, to investigate the 3-D shape of human adenovirus using a technique called X-ray crystallography.
First the researchers needed to grow a crystal of the virus. This presented several challenges. For one, a complete human adenovirus sports fiber "spikes" that help it attach to a host cell, but also prevented researchers from successfully growing a crystal. One of the first steps the scientists took was to engineer a human adenovirus molecule without the fiber spikes. Altogether, getting usable crystals took five years.
When they were ready to test the virus crystal, Nemerow and Reddy partnered with Argonne biophysicist Robert Fischetti and started their work at the Advanced Photon Source's General Medicine and Cancer Institute's Collaborative Access Team (GM/CA) beamline.
Unlike other beamlines they'd tested, the GM/CA beamlines produce intense X-ray beams that allowed the researchers to collect high-quality data from the virus crystals. This gave researchers a detailed 3-D image of the structure—the largest structure of any biomolecule determined by X-ray crystallography to date.
"Without the beamline, we would have been stymied on this project," said Nemerow. The APS beamline, they hoped, would reveal where electrons were clustered in the human adenovirus molecule and how they were connected.
Over the course of the project, Fischetti and colleagues modified the beamline to improve the uniformity of the beam. They installed smoother mirrors, reduced the amount of radiation damage to the crystals, and provided the team more flexibility to focus the beam. This helped Nemerow and Reddy hone the first electron density map of the human adenovirus' structure, which they published in the journal Science.
Nemerow and Reddy compared their map to what was already known about the human adenovirus from previous research. "We weren't able to assign an identity for all 12 of the proteins in human adenovirus to the electron density we mapped out," Nemerow said. "There were also four major 'cement' proteins we could not identify or assign—either because they haven't been seen yet, were too disordered or because we didn't have enough data at the time."
Cement proteins, as their name suggests, bind major proteins to the virus shell, or capsid.
Nemerow and Reddy found major capsid proteins tended to be easier to see in an electron density map because of their symmetry and general stability, but cement proteins proved the sticking point. "We don't know the structures of cement proteins at high resolution yet, because they are difficult to purify by themselves—which makes them even harder to crystallize," Reddy explained. "The cement proteins bind to major capsid proteins, like cement to bricks, and mediate interactions between them." Because cement proteins are flexible and dynamic, they can degrade easily from radiation damage or fail to produce diffraction under a high-resolution X-ray.
Fischetti and colleagues are upgrading the GM-CA beamlines to provide smaller and more intense X-ray beams to help scientists create clearer images from their crystals. Once the upgrade is completed, Nemerow and Reddy can continue to complete their map of the human adenovirus, so that one day a seasonal nuisance may become a powerful tool for human health.
Journal information: Science
Provided by Argonne National Laboratory