September 20, 2012 feature
Small is beautiful: Viewing hydrogen atoms with neutron protein crystallography
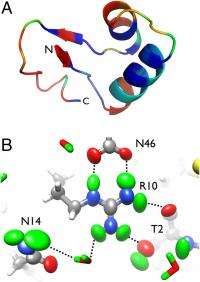
(Phys.org)—Creating 3D visualizations of hydrogen atoms in proteins is especially challenging, often requiring their locations to be inferred from those of nearby carbon, nitrogen, oxygen or sulfur atoms stored in protein structure databases. These locations are based on atomic positions in databases of previously solved structures, general chemical knowledge, quantum mechanical calculations, or potential hydrogen bonding interactions. While X-ray crystallography can pinpoint hydrogen atom locations at ultrahigh resolution, in practice only a few such positions are experimentally determined. Recently, however, scientists at the University of Toledo, Los Alamos National Laboratory and Oak Ridge National Laboratory used neutron crystallography – a technique that even at lower resolutions can locate individual hydrogen atoms by leveraging scattering properties of the hydrogen isotope deuterium.
Research Professor Julian C.-H. Chen discussed the group's research with Phys.org, first summarizing the main challenges they faced in overcoming the limitations of only X-ray crystallography, visualizing hydrogen atoms a three-dimensional context, determining protonation states of active site residues in enzymes, and establishing the identities and orientation of solvent molecules. "The strength of neutron crystallography lies in its ability to locate hydrogen atoms in macromolecular structures. Hydrogen accounts for one-half of the atoms in any given protein, and help perform all sorts of tasks, from hydrogen bonding to catalysis," Chen explains. "For X-ray crystallography, locating hydrogen atoms is especially difficult as they possess just one electron, and X-rays are weakly scattered by hydrogen atoms compared to other elements in proteins such as carbon, oxygen, nitrogen, and sulfur."
Furthermore, hydrogen atoms in protein structures can be quite mobile. "You're lucky to be able to see them even in the highest resolution structures," Chen continues. "Neutrons, on the other hand, have the nifty property that deuterium, which can be introduced into reactive sites on the protein through hydrogen/deuterium exchange, scatters neutrons so they are easily located and readily visible in nuclear density maps, even at moderate resolution. So this makes it relatively straightforward to determine protonation states of residues. Similarly, for solvent molecules, D2O, or heavy water, can be introduced into the crystal – and these molecules have an easily identifiable boomerang shape in the nuclear density maps, which makes it possible to orient them."
On addressing these challenges, Chen adds, "We reasoned that if we had the combination of ultrahigh resolution and neutron diffraction data on hydrogen/deuterium-exchanged crystals, we should be able to describe how individual deuterium atoms vibrate – and not only that, but also describe their anisotropic vibrational motion." (An anisotropic material exhibits different values of a property in different crystallographic directions.) "So that's exactly what we did for a number of selected deuterium atoms within crambin – a protein that forms the best-ordered macromolecular crystals known. We surmised that crambin crystals would diffract neutrons to ultra-high resolution as well."
As Chen points out, crambin is a small protein with a limited number of deuterium atoms. "Especially with the new neutron crystallography instruments under construction or being commissioned, new datasets of similar or higher resolution should be possible with crambin as well as other proteins. We're of course staying updated on these developments." Chen adds that some of their findings pertaining to hydrogen/deuterium exchange can also be addressed by other physical methods.
Chen also comments on their key findings provide the ability to inventory all hydrogen bonding interactions within the protein and ascertain the categories and suggest strengths of hydrogen bonds in the system; demonstrate the power of ultrahigh resolution neutron diffraction to resolve ambiguous hydrogen bonding networks; and reveal unconventional structural features and interatomic interactions. "You generally only see the oxygen of the water in X-ray structures, keep in mind that the two hydrogens scatter X-rays weakly and are generally invisible." This means that the hydrogen bonding networks can be ambiguous. "But with neutrons," Chen continues, "since you get strong scattering from the deuteriums of the D2O, you can orient the hydrogens and figure out the network of hydrogen bond donors and acceptors."
Moreover, says Chen, the researchers also saw some instances of unusual hydrogen/deuterium exchange patterns. "One residue in particular appeared to have hydrogen/deuterium exchange on one of the hydrogens bonded to the alpha carbon. This could be evidence of C-H. . .O hydrogen bonds, which are rather elusive and difficult to observe experimentally although nearly 30 years of mostly computational work have been done on these hydrogen bonds."
In terms of other research and downstream technology might benefit from your findings, Chen stresses that their study demonstrates what can be done with neutron crystallography. "I think that this, and future structures, will give the humble hydrogen atom its due," Chen concludes. "This will hopefully work its way into computational algorithms that address protein-ligand and protein-drug interactions. This work may also lead in the long run to more accurate, higher quality macromolecular structures."
More information: Direct observation of hydrogen atom dynamics and interactions by ultrahigh resolution neutron protein crystallography, PNAS September 4, 2012, doi:10.1073/pnas.1208341109
Journal information: Proceedings of the National Academy of Sciences
Copyright 2012 Phys.org
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.