Development of antibiotic resistance more predictable than expected
Research by Wageningen University has shown that the development of bacteria with resistance against the antibiotic cefotaxime occurs more often and more predictably than was previously assumed. Bacterial populations were found to have many mutations that increase resistance and therefore have a negative effect on public health. Moreover, the effects are such that it can be predicted that the development of bacterial strains with a resistance against cefotaxime will progress in a similar way in different patients from different locations. Together with German colleagues, the Wageningen scientists developed a research approach which will allow them to predict whether, and if so how, resistant bacterial strains will develop for other antibiotics as well.
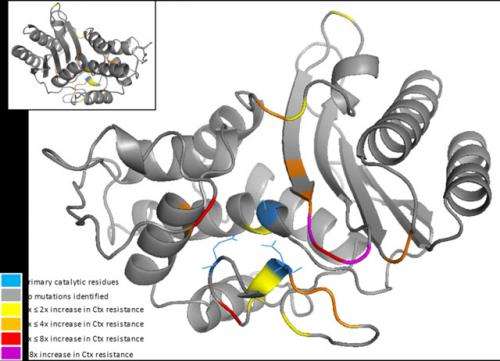
The Wageningen scientists studied the main enzyme that causes resistance against the antibiotic cefotaxime. The only function of this beta-lactamase enzyme is the breakdown of so-called beta-lactam antibiotics, which kill bacteria by preventing the production of their cell walls. Martijn Schenk and Arjan de Visser, genetic scientists at Wageningen University, were surprised by the number of mutations with a positive effect on the resistance against cefotaxime.
De Visser: “Of all the mutations we found in this beta-lactamase, more than three per cent caused an increase in the resistance against the antibiotic. To top it all off, we discovered that the mutations with a strong effect also had a much greater impact than we had anticipated. Based on theoretical arguments and previous observations, we had estimated the effects on the resistance against the antibiotic to be significantly lower.”
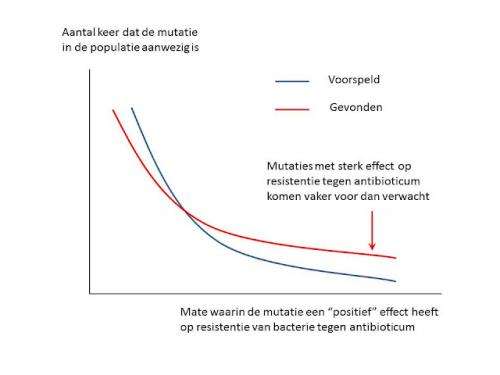
The presence in particular of mutations with a very strong effect on resistance to the antibiotic facilitates the prediction of the development of resistant bacterial strains.
Collaboration with a group of physicists in Germany enabled the Wageningen scientists to study the genetic findings quantitatively, as Martijn Schenk explains: “The physicists built computer models that helped us as geneticists to move forward. We were able to show that it is probable that the bacteria will become resistant against the antibiotic in a similar way in various patients throughout the world.”
According to De Visser the approach taken can also be used to predict the ‘tenability’ of other antibiotics, as the combination of computer models with knowledge about the number and effect of the mutations provides concrete leads.
The research results have been published in PLoS Genetics last week. The research was partly financed by the Deutsche Forschungsgemeinschaft (DFG), Germany's largest research funding organisation.
More information: Schenk, MF, IG Szendro, J Krug and JAGM de Visser. 2012. Quantifying the adaptive potential of an antibiotic resistance enzyme. PLoS Genetics 8(6): e1002783
Journal information: PLoS Genetics
Provided by Wageningen University