Channeling our ion past

To understand how the first cells transported ions across their membranes, researchers are studying simple channels used by fungi to kill off bacteria.
The ability to shuffle ions in and out of a cell is clearly vital to life as we know it, but how the necessary membrane channels originated is a mystery. A new research project is exploring some simple protein-like structures in fungi that may point the way to the first ion channels.
Modern cells have semi-permeable membranes that largely prevent material from passing through. But a cell still needs to bring in food and get rid of waste. For this reason, life has evolved complex molecular structures that essentially pry open a hole in the membrane.
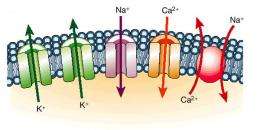
Ion channels are protein assemblies that provide passage through a membrane They let ions like sodium and potassium go either in or out of a cell, depending on which direction the lower concentration is. This sort of charge transport is needed for a variety of tasks such as regulating the volume of the cell, sensing the environment, stabilizing DNA molecules and generating chemical energy in cells.
On larger scales, the beating of our heart and the electric hum inside our brain rely heavily on the action of ion channels.
"We wouldn't be having this discussion if our ion channels weren't working properly," says Andrew Pohorille of NASA’s Ames Research Center.
It is evident that life has relied on ion channels for a long time, but the current forms of these protein assemblies are extremely complicated.
"What's their history? Where did they come from?" Pohorille wonders. "At their origin, they undoubtedly were simpler than they are now."
Pohorille and his colleagues are trying to piece together a plausible backstory for ion channels. They are focusing on self-assembling molecular complexes that some fungi use to combat bacteria. Previous work has shown that these structures can function as rudimentary ion channels.
With funding from the NASA Exobiology and Evolutionary Biology program, Pohorille's team will be looking to see how these simple "tunnels" could have developed the ability to regulate the flow of ions through them.
Drawing borders
The very first biological cells may have succeeded without ion channels and other similar structures. If their membranes were "leaky," then nutrients and waste could have flown in and out without special openings.
Dave Deamer of UC Santa Cruz has studied this possibility. He claims lipid bilayers may have provided a self-assembling membrane for the first life forms. These "soapy films" are more permeable than current cell membranes made from phospholipids.
However, Deamer admits that these simple membranes probably aren't permeable enough. He and others have done experiments trying to pass certain biomolecules through a lipid bilayer.
"It worked, but we had to wait several days," Deamer says.
Pohorille thinks "leaky" is the wrong strategy for biology.
"The problem is that this situation eventually leads nowhere," Pohorille says. "You only get serious about the evolution of life when a cell retains the things that it makes."
Ion channels were likely one of the first type of membrane-spanning molecular assemblies. These channels are passive, which means that they rely on the natural diffusion of ions from, say, a high concentration inside a cell to a low concentration outside. This distinguishes them from ion pumps (a presumably later evolutionary development) that use energy to drag ions in a particular direction.
But modern ion channels are not simple "holes in the wall." Each channel is composed of around 5 to 10 very large proteins, which intricately work together to open and close the channel when necessary.
"These are some of the most complicated structures in the cell," Pohorille says.
Recently it was discovered that microbes rely on ion channels that resemble by and large those in complex organisms.
"It came as a surprise, since microbes don't have a heart or a nervous system," Pohorille says.
But one important difference is that the proteins in microbial channels are easier to analyze than those in higher organisms. Biologists have therefore made a lot of progress in understanding how these structures work.
Channels of death
One of the remaining questions about ion channels concerns their origin. Could the first ion channels have formed without all the complex protein machinery of a modern cell?
As it turns out, biology does provide a possible prototype of a primitive ion channel, but it comes from a somewhat unexpected place: a bacteria-fighting mechanism found in fungi. It relies on "peptaibols," which are protein-like molecules that readily come together to form tubes. The fungi use them to puncture a hole in an invading bacteria and kill it by letting the insides leak out.
What makes these peptaibols special is that they contain non-standard amino acids that normal proteins do not. This unique chemistry is what makes the peptaibol-based "spears" so deadly. The bacteria has enzymes for breaking down proteins, but none of these enzymes are tailored for a molecule made up of non-standard amino acids.
Because DNA has no code for non-standard amino acids, the fungi have a separate mechanism that builds each peptaibol up, one amino acid at a time.
"It's a painfully slow procedure, so the structures can't be very long," Pohorille says.
Since these molecules aren't subject to the evolutionary effects of DNA mutations, they may be better preserved than other biomolecules. Thus, they may harken back to earlier times in our evolutionary past.
"We think they are very good models of what the first ion channels might have looked like," Pohorille says.
Experiments have shown that the peptaibol channels are remarkably efficient at transferring ions. And these channel properties are quite robust. Tests show that different arrangements of the constituent amino acids still gives a working channel. Such non-specificity would presumably have been an asset to the earliest channels, which likely formed without a DNA blueprint.
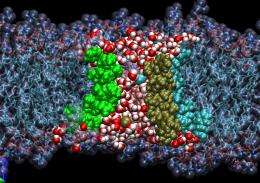
Pohorille's team has developed computer simulations of peptaibol channels. In their models, they embed a channel in a phospholipid membrane and place ions in the surrounding fluid.
The researchers have been surprised how flexible, or floppy, the channels are in these simulations. The peptaibol structures are moving around so much that it doesn't appear as if they really offer an open passageway. But, on average, ions are getting through quite well, Pohorille says.
Normal ion channels, by contrast, are fairly rigid. This rigidity presumably makes them more predictable.
Pohorille thinks this may be pointing to a larger trend. Early proteins may have started out being very flexible. Only later, after many generations of natural selection, did these structures become rigid and thus more precise in their function.
An open-close switch
One thing that the simple peptaibol-based channels lack is the ability to open and close "on command." This sort of regulation requires sophisticated machinery in order to respond to some environmental stimulus. For example, voltage-gated ion channels open and close in response to a membrane potential. Other channels are triggered by the presence of a specific molecule or ion.
Pohorille and his team are currently working on how regulation machinery could be added to a peptaibol channel.
Recently, a Japanese group found they could regulate the activity of one of these simple channels by the adding iron atoms with different oxidation states. Pohorille believes it shouldn't be too hard to go from this to a channel that would be regulated by pH or the presence of some small molecule.
"This would be a bridge to a contemporary channel," Pohorille says.
Deamer, who is not involved in this project, thinks this research is complementary to other work that is trying to synthesize a working cell in the lab.
"Pohorille is adding a piece in the pathway to artificial life," Deamer says.
He says that a better understanding of ion channels could help guide his own work studying the first membranes.
Source: Astrobio.net