Bacteria to the rescue
At several U.S. Department of Energy (DOE) sites, uranium mining, milling, and processing have led to groundwater contamination that persists above drinking water standards—in spite of natural flushing and the removal of contaminated sediments. A multi-institutional research team, including researchers at DOE's Pacific Northwest National Laboratory (PNNL), is investigating the use of bioremediation to treat the subsurface uranium plumes and remove pollutants. Research conducted by the team at the Integrated Field Research Challenge (IFRC) site in Rifle, Colorado has shown that indigenous bacteria can be stimulated to immobilize the uranium, resulting in groundwater concentrations below the U.S. Environmental Protection Agency standard.
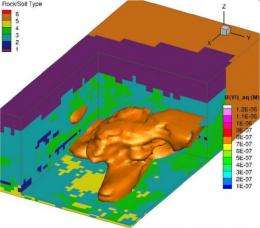
A key component of the research is the use of high-performance computers and comprehensively detailed simulation codes to better understand the subsurface processes, properties, and conditions controlling uranium behavior. Using a code designed to efficiently exploit the large memory and high-performance of advanced computers, PNNL researchers were able to incorporate more processes and interactions of interest at higher levels of detail, with simulations completing in hours rather than weeks.
To account for the high spatial and temporal resolution, and the large number of reactive species, minerals, and microbially mediated reactions, researchers used PNNL's eSTOMP subsurface simulator on the Environmental Molecular Sciences Laboratory's (EMSL's) massively parallel supercomputer, Chinook, to model the 110-day in situ field experiment and 50 days of post-biostimulation behavior. The capability included variably saturated flow and biogeochemical reactive transport through three-dimensional physically and chemically heterogeneous sediments, as well as new knowledge about the behavior and interaction of the stimulated microbial community with the subsurface geochemical environment. In addition to the metal-reducing bacteria that catalyze the formation of immobile uranium, researchers demonstrated the importance of accounting for sulfate-reducing bacteria activity-finding that uranium bioreduction is most effective when acetate concentrations are engineered to exceed the sulfate-reducing bacteria demand.
Researchers are working to incorporate genome-scale metabolic models into eSTOMP field-scale simulations to more accurately account for the biologically-mediated reactions and rates. The approach builds on advancements in the collection and analysis of proteomic data that are used to assess and validate the large number of detailed cellular process mechanisms in naturally complex field settings.
More information: Yabusaki SB, et al. 2011. "Variably saturated flow and multicomponent biogeochemical reactive transport modeling of a uranium bioremediation field experiment." Journal of Contaminant Hydrology 126(3-4):271-290. DOI:10.1016/j.jconhyd.2011.09.002
Provided by Pacific Northwest National Laboratory