Microprinting Technique for Patterning Single Molecules
A new process for creating patterns of individual molecules on a surface combines control of self-assembled monolayers (SAMs) and a soft lithography technique known as microcontact printing. Scientists use the process, known as "microcontact insertion printing" to build surfaces that have molecules with specific functions inserted at known intervals on a surface.
The new technique, with potential applications ranging from analysis of biochemical mixtures to molecular-scale electronic components, will be described as the cover story of the 5 February 2007 issue of the journal Applied Physics Letters by a team led by Penn State researchers Paul S. Weiss, distinguished professor of chemistry and physics; Mark Horn, associate professor of engineering science and mechanics; and Anne M. Andrews, assistant professor of veterinary and biomedical sciences.
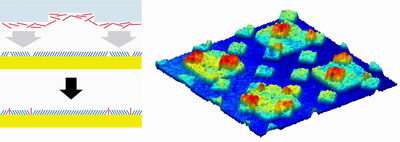
Microcontact insertion is based on the technique of microcontact printing, in which a patterned rubber-like stamp is "inked" with a solution of molecules and then applied to a surface. However, the insertion technique does not apply molecules to the entire surface contacted, but instead fills only defects -- molecule-sized gaps -- in a layer of molecules that previously has been placed on and attached to the surface. "Lithography cannot place molecules with nanometer precision," says Weiss, "but by building the defects into the surface and then filling them selectively with this process, we can place the isolated molecules in a predesigned nano-scale or micro-scale pattern."
The process of microcontact insertion printing starts with a self-assembled monolayer (SAM) -- a chemical deposition on the surface that is one molecule thick. The researchers can build the SAM with defects, or regions in which the surface is not covered by the film. By controlling the type, size, and number of the defects, they create a pattern in which the surface appears as a matrix of exposed dots. In one example, there is an average of 10 molecules between defects, making an average separation between inserted molecules of about 5 nanometers. If defects are made larger, then more molecules are inserted in each one.
After the formation of the matrix with controlled defects, the microcontact printing technique is used to fill the exposed parts of the surface. "We use the stamp to attach molecules to the open surface," says Weiss. "Because each molecule is surrounded by the SAM, it stays in place and there is no migration." Using a series of stamps allows different molecules to be placed on the surface in a pattern, with each region of the surface holding a different type of functional molecule tethered to the surface and held in place by the surrounding inert monolayer.
The chemical functionality of the attached molecule can be made in a way that will capture specific types of molecules from a mixture. The pattern of functionalities creates a multiplexed capture surface on which chemical compounds, such as proteins, other biological molecules, or environmental contaminants can be separated from a complex mixture. Each part of the pattern can be designed for specific classes of target molecules so that a single patterned surface can be used to determine the identity and concentration of multiple components of the mixture.
In addition to its application in creation of surfaces with specific selectivity, the microcontact-insertion process could allow the controlled deposition of molecules that can interact in specific ways. This application could be used to build electronic components or other functional surfaces.
Source: Penn State