November 14, 2014 weblog
Researchers build a map of sites where genetic information is swapped between chromosomes

Bob Yirka
news contributor
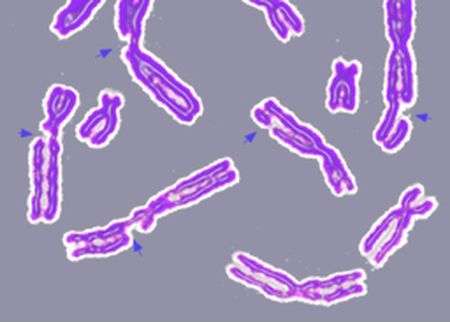
(Phys.org)—A small team of researchers in the U.S. has built a detailed map that shows the sites where human genetic information is swapped between chromosomes. In their paper published in the journal Science, the team describe their work and explain how their maps could provide new ways of looking at how such sites influence evolution and possibly genetic diseases. Bernard de Massy, of the Institute of Human Genetics offers an in depth perspective piece on the work done by the team in the same journal edition.
In order to reproduce, the human body must first make either sperm in males or eggs in females and to do that, it must somehow create cells that have just half of that individual's DNA—the other half will come from the other person during conception. The process by which this occurs is called homologous recombination, where diploid cells undergo cell division resulting in the production of gamete cells. The process is initiated when the body forms breaks in DNA double-strands. It's in the repair of these breaks that exchanges between chromosomes occur. Recombination occurs at specific sites along chromosomes that are known as hot spots. Finding these hot spots has proved challenging, but now, the researchers with this latest effort report that they have succeeded in creating map of such hot spots which not only allows for looking at how evolution works in new ways, but could prove to be very helpful in better understanding, and possibly treating, some genetic diseases.
In their study, the researchers were able to detect the triggers inside the body that led to recombination occurring in human male volunteers—that allowed them to create the maps which show where the recombination events occur. Just looking at the maps, the team notes, allows for a better understanding of the recombination process. One preliminary finding, for example, was that hotspots can be influenced by proteins expressed by a specific gene. de Massy notes that the maps created by the team could very well be used in other mammal species as well, which could help researchers studying how genetic diseases come about better understand where things go wrong, and perhaps, shine a light on possible ways to prevent it from happening.
Written for you by our author Bob Yirka—this article is the result of careful human work. We rely on readers like you to keep independent science journalism alive. If this reporting matters to you, please consider a donation (especially monthly). You'll get an ad-free account as a thank-you.
Publication details
Recombination initiation maps of individual human genomes, Science 14 November 2014: Vol. 346 no. 6211. DOI: 10.1126/science.1256442
ABSTRACT
DNA double-strand breaks (DSBs) are introduced in meiosis to initiate recombination and generate crossovers, the reciprocal exchanges of genetic material between parental chromosomes. Here, we present high-resolution maps of meiotic DSBs in individual human genomes. Comparing DSB maps between individuals shows that along with DNA binding by PRDM9, additional factors may dictate the efficiency of DSB formation. We find evidence for both GC-biased gene conversion and mutagenesis around meiotic DSB hotspots, while frequent colocalization of DSB hotspots with chromosome rearrangement breakpoints implicates the aberrant repair of meiotic DSBs in genomic disorders. Furthermore, our data indicate that DSB frequency is a major determinant of crossover rate. These maps provide new insights into the regulation of meiotic recombination and the impact of meiotic recombination on genome function.
Journal information: Science
© 2014 Phys.org