April 18, 2012 feature
Adam's rib, revisited: Evolutionary divergence of mammalian sex chromosomes
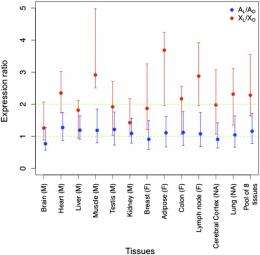
(Phys.org)—Males and females... Mars and Venus... XY and XX chromosomes—all are common memes. At the same time, the evolution of therian (placental and marsupial) sex chromosomes is less widely understood. More to the point, these arose some 150 million years ago from a pair of autosomes, or non-sex chromosomes. Having appeared, the X and Y chromosomes – both with the same ancestral genes – began diverging, with the Y chromosome evolving into a state in which (except for two small autosomal regions) it never recombines. As a result, the Y chromosome has degenerated, losing most of its genes in the process. On the other hand, the X chromosome does recombine, retains many ancestral genes – and has gained new genes, and evolved new expression patterns, as well.
The increased imbalance of X/Y chromosomal loci led to the emergence of loci-specific X chromosome inactivation, which has been seen as compensating for differential gene dosage (the number of copies of a given gene present in a cell or nucleus) by making expression of X-linked genes similar in males and females. Recently, using RNA sequencing, or RNA-seq, data (more precise than previously-analyzed microarray data), scientists in the Laboratoire de Biométrie et Biologie Évolutive, Université Lyon, Centre National de la Recherche Scientifique, in Villeurbanne, France, found support for the hypothesis that XCI acts as a dosage-compensation mechanism. At the same time, the scientists explored the contribution of dosage-sensitive genes to phenotype expression in X aneuploidy – a condition, relatively common in humans, in which one or more extra or missing chromosomes leads to an unbalanced chromosome complement, resulting in conditions such as Turner (X0) and Klinefelter (XXY) syndromes.
Associate Professor Gabriel A. B. Marais, PhD student Eugénie Pessia and other researchers faced a number of challenges in designing and implementing research to determine how and why female somatic X-chromosome inactivation evolved in mammals – especially given that it has been poorly understood. "When X-chromosome inactivation, or XCI, was first described in the 1960s, Susumu Ohno suggested that XCI evolved as part of a dosage compensation mechanism involving X chromosome hyperexpression in both sexes coupled with inactivation of one of the X chromosomes in females," Marais tells Phys.org. "However, the X hyperexpression part of Ohno's scenario has remained completely speculative for a very long time. Only recently, several studies have tried to compare the global X versus autosomal expression – the so-called X/A expression ratio – using microarray or RNA-seq data. Some found an X/A close to 1, others close to 0.5 – but the most recent studies from 2011 found a ratio of 0.7, which was difficult to interpret. We thought that maybe only some of X-linked genes need to be hypertranscribed and dosage-compensated, which would explain the X/A of 0.7 when all X-linked genes – that is, some with an X/A of 1 and some with an X/A of 0.5 – are analyzed together. We thought that it would make perfect sense if only the dosage-sensitive genes on the X chromosome would have an X/A of 1, as they're the ones that need a precise dosage."
The research team addressed these issues by studying gene expression of dosage-sensitive genes on the X chromosome – but since those are not very well known. "Based on studies in plants, yeasts and also in humans, we thought we should focus on protein-complex genes," Marais explains. "Protein complexes require a precise stochiometry to be functional, which means that if one protein is much more abundant than the other proteins of the complex, it is likely to be nonfunctional, which may be harmful for the cell." They used the data on human complexes from the HPRD database to get our list of protein-complex genes on the X chromosome.
"For large complexes," Marais continues, "we found that the expression levels estimated by RNA-seq are similar for X-linked and autosomal genes of a same complex. This is quite striking because both in male and female, only one X is present or expressed, whereas both autosomes are expressed. The X-linked genes have only one expressed copy and the autosomal two, so the expression of the X-linked genes should be half that of the autosomal ones, but we found it to be the same, which means the X-linked genes are hyperexpressed. How this hyperexpression is achieved remains an open question, but recent data in mice suggests that RNA polymerase II could be more abundant on these X-linked genes, probably because of epigenetic marks."
Looking ahead, Marais notes, "We'd like to study the other dosage-sensitive genes on the X chromosome, as we know that protein-complex genes are not the only ones. Genes involved in regulatory networks are probably dosage-sensitive. However, we need better characterization of these networks in humans before extending our analysis to these other dosage-sensitive genes."
Marais adds that the team would like to understand how XCI originated. "We showed that XCI is part of a dosage compensation mechanism for dosage-sensitive X-linked genes, because XCI downregulates their expression to get a balanced X and autosomal expression, and thereby avoid an X/A of 2, in females. However," he acknowledges, "our study does not provide insights on how XCI evolved in the first place. X-inactivation is found in placentals and marsupials – and in marsupials and in some tissues of some placentals, it is the paternal X that is always inactivated. David Haig has proposed that X-inactivation might originally be a form of genetic imprinting, which evolved because of parental conflicts. We'd like to test this idea."
Marais also points out that it might well be possible to transition to in silico modeling. "Actually, there's very little theoretical work on the evolution of dosage compensation. The hypothesis proposed by David Haig has not been modeled at all," he notes, "and it would be important to show by simulation that his scenario can work."
Regarding research, technology and applications might benefit from their findings, Marais notes that in humans, most aneuploidies (aberrant number of chromosomes) are lethal. "Trisomy 21 is an exception, but there are strong effects on the phenotype known as Down syndrome. The X chromosome is another exception, and X aneuploidies have surprisingly mild effects given the size of this chromosome and the number of genes it contains compared to chromosome 21 – one of the smallest chromosomes in our genome. This is because all the supernumerary X chromosomes are inactivated by XCI, so in XXY individuals or X0 individuals, only one X is expressed just like in XX or XY individuals. However, about 15% of the genes located on the human X chromosome escape XCI, and for these ones X aneuploidies will have consequences on gene dosage. Our results suggest that the consequences will be stronger for the dosage-sensitive genes." Marais concludes. He suggests that genes being both dosage-sensitive and XCI-escapees are probably the best candidates for underlying X aneuploidy syndromes such as Turner (X0), Klinefelter (XXY) and Triple-X. "The identification and characterization of such genes would of course help develop treatments that some patients affected by these very frequent syndromes need."
More information: Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome, Published online before print March 5 2012, PNAS April 3 2012 vol. 109 no. 14 5346-5351, doi: 10.1073/pnas.1116763109
Copyright 2012 Phys.Org
All rights reserved. This material may not be published, broadcast, rewritten or redistributed in whole or part without the express written permission of PhysOrg.com.















