Faster, cheaper DNA sequencing method developed
(PhysOrg.com) -- Boston University biomedical engineers have devised a method for making future genome sequencing faster and cheaper by dramatically reducing the amount of DNA required, thus eliminating the expensive, time-consuming and error-prone step of DNA amplification.
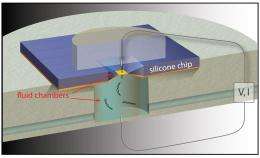
In a study published in the Dec. 20 online edition of Nature Nanotechnology, a team led by Boston University Biomedical Engineering Associate Professor Amit Meller details pioneering work in detecting DNA molecules as they pass through silicon nanopores. The technique uses electrical fields to feed long strands of DNA through four-nanometer-wide pores, much like threading a needle. The method uses sensitive electrical current measurements to detect single DNA molecules as they pass through the nanopores.
"The current study shows that we can detect a much smaller amount of DNA sample than previously reported," said Meller. "When people start to implement genome sequencing or genome profiling using nanopores, they could use our nanopore capture approach to greatly reduce the number of copies used in those measurements."
Currently, genome sequencing utilizes DNA amplification to make billions of molecular copies in order to produce a sample large enough to be analyzed. In addition to the time and cost DNA amplification entails, some of the molecules - like photocopies of photocopies - come out less than perfect. Meller and his colleagues at BU, New York University and Bar-Ilan University in Israel have harnessed electrical fields surrounding the mouths of the nanopores to attract long, negatively charged strands of DNA and slide them through the nanopore where the DNA sequence can be detected. Since the DNA is drawn to the nanopores from a distance, far fewer copies of the molecule are needed.
Before creating this new method, the team had to develop an understanding of electro-physics at the nanoscale, where the rules that govern the larger world don't necessarily apply. They made a counterintuitive discovery: the longer the DNA strand, the more quickly it found the pore opening.
"That's really surprising," Meller said. "You'd expect that if you have a longer 'spaghetti,' then finding the end would be much harder. At the same time this discovery means that the nanopore system is optimized for the detection of long DNA strands -- tens of thousands basepairs, or even more. This could dramatically speed future genomic sequencing by allowing analysis of a long DNA strand in one swipe, rather than having to assemble results from many short snippets.
"DNA amplification technologies limit DNA molecule length to under a thousand basepairs," Meller added. "Because our method avoids amplification, it not only reduces the cost, time and error rate of DNA replication techniques, but also enables the analysis of very long strands of DNA, much longer than current limitations."
With this knowledge in hand, Meller and his team set out to optimize the effect. They used salt gradients to alter the electrical field around the pores, which increased the rate at which DNA molecules were captured and shortened the lag time between molecules, thus reducing the quantity of DNA needed for accurate measurements. Rather than floating around until they happened upon a nanopore, DNA strands were funneled into the openings.
By boosting capture rates by a few orders of magnitude, and reducing the volume of the sample chamber the researchers reduced the number of DNA molecules required by a factor of 10,000 - from about 1 billion sample molecules to 100,000.
More information: The article, "Electrostatic Focusing of Unlabelled DNA into Nanoscale Pores Using a Salt Gradient," is available at the Nature web site at dx.doi.org/10.1038/natureNNANO.2009.379
Provided by Boston University
















