Biologists Discover How 'Silent' Mutations Influence Protein Production
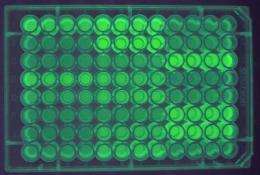
(PhysOrg.com) -- Biologists at the University of Pennsylvania have revealed a hidden code that determines the expression level of a gene, providing a way to distinguish efficient genes from inefficient ones. The new research, which involved creating hundreds of synthetic green-glowing genes, provides an explanation for how a cell "knows" how much of each protein to make, providing just the right amount of protein to maintain homeostasis yet not too much to cause cell toxicity.
In the study, Penn biologists analyzed how protein levels are governed by synonymous, or silent, mutations within the protein-coding region.
Synonymous mutations do not change the amino-acid sequence of a protein, but they can nevertheless influence the amount of the protein that is produced. The researchers identified the mechanism underlying this regulation: synonymous mutations determine mRNA folding and thereby the eventual protein level. The researchers also identified a class of mutations that did not directly affect protein levels but slowed bacterial growth.
For biologists, these results fundamentally change the understanding of the role of synonymous mutations, which were previously considered evolutionarily neutral. The findings may also improve the design of therapeutic genes. Many drugs, such as insulin, are produced by transgenic cell lines. Using optimized genes will produce larger amounts of therapeutic proteins while keeping the transgenic, carrier cells healthy and fast-growing.
The human genome contains more than 20,000 genes that encode the proteins present in a human body. Some of these proteins are needed in bulk, while for others a tiny amount is sufficient and a large amount would be toxic. The question is how cells "know" how much of each protein to make.
To answer this question, Joshua B. Plotkin, senior author and the Martin Meyerson Assistant Professor in the Department of Biology in Penn's School of Arts and Sciences, and colleagues at Harvard University and the University of Edinburgh engineered a synthetic library of 154 genes that varied randomly at synonymous sites. All the genes encoded the same green fluorescent protein, enabling the researchers to easily study the effects of such mutations on protein levels when expressed in the bacterium Escherichia coli.
The silent mutations changed the amount of fluorescent protein by as much as 250-fold, without changing the properties of the protein. Codon bias, the probability that one codon of three adjacent nucleotides will code for one amino acid over another, was previously thought to be the cause for protein expression variance, but it did not correlate with gene expression in these experiments.
"At first we were stumped," Plotkin said. "How were the silent mutations influencing protein levels? Eventually, we looked at mRNA structure and discovered that this was the underlying mechanism."
The stability of mRNA folding near the ribosomal binding site explained more than half the variation in protein levels. To understand this observation, the researchers simulated the spatial arrangement of the messenger RNA molecule that carries the information from genes to proteins. They found that the inefficient genes produced tightly folded mRNA molecules that could not be accessed by the protein-making machinery. According to their analysis, mRNA folding and associated rates of translation initiation play a predominant role in shaping expression levels of individual genes, whereas codon bias influences global translation efficiency and cellular fitness.
The study, appearing in the current issue of the journal Science, was performed by Plotkin, as well as first author Grzegorz Kudla of the Department of Biology at Penn, Andrew W. Murray of the Department of Molecular and Cellular Biology at Harvard and David Tollervey of the Wellcome Trust Centre for Cell Biology at Edinburgh.
Source: University of Pennsylvania















