Scientists discover mobile small RNAs that set up leaf patterning in plants
A key item in the developmental agenda of a plant leaf is the establishment of an axis that makes a leaf's top half distinct from its bottom half. This asymmetry is crucial for the leaf's function: it ensures that the leaf develops a flattened blade that is optimized for photosynthesis, with a top surface specialized for light harvesting and a bottom surface containing tiny pores that serve as locales for gas exchange.
For years, plant biologists have known that this top/bottom axis - analogous to the front/back or "dorso-ventral" axis in animals - is established by a signal derived from the meristem, the stem cell-rich growing tip of the plant from which new leaves arise. Other signals that traffic between the upper and lower sides of the leaf are thought to stably maintain this polar axis. Just as a GPS signal tells drivers where they are, these signals give cells positional information about where they are located within the leaf, causing them to acquire their correct identities by switching specific genes "on" or "off."
Associate Professor Marja Timmermans, Ph.D., and her team of scientists at Cold Spring Harbor Laboratory (CSHL) are the first group to uncover the identity of one such positional signal. In a study that appears in the March 1st issue of Genes and Development, they describe a family of mobile small RNAs that patterns the top/bottom axis in leaves. These small RNAs, they discovered, are generated on the upper surface of young leaves but traffic from this source to form a concentration gradient across each leaf. This graded distribution pattern creates discrete regions of gene activity so that cells in each half of a leaf develop a distinct "top" or "bottom" identity.
"We've known that small RNAs produced upon viral infection can move from cell to cell," explains Timmermans. "But this is the first time anyone has shown that small RNAs that are native to the organism are similarly mobile and set up developmental patterns when they move through neighboring cells."
Ta-siRNAs behave like morphogens
These native, or endogenous, small RNAs are called trans-acting small interfering RNAs (ta-siRNAs). Like microRNAs and endogenous siRNAs present in other organisms, they regulate gene activity via a mechanism called RNA interference. Because of their newly discovered properties in leaf patterning, Timmermans likens the ta-siRNAs to "morphogens" or form-generating substances. Morphogens have been well studied in animals, although to date, scientists have only discovered protein and hormone morphogens. These molecules operate as positional signals whose effect on target cells is concentration-dependent. Secreted at a defined location, their movement establishes a concentration gradient that patterns a developing tissue such that cells closest to the morphogen's point of origin become distinct from cells that are farther away.
The CSHL team has now found that in plant leaves, ta-siRNAs similarly generate a concentration gradient that divides the developing leaf into a top and bottom half with different specialized cell types.
A ta-siRNA 'gradient' determines the ups and downs of developing leaves
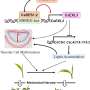
The function of these ta-siRNAs is to specifically block the activity of a gene called ARF3. This gene defines the identities of cells found in the bottom half of leaves. For the correct leaf pattern to develop, it is therefore crucial that ARF3 is switched "on" in the right cells - those at the leaf's lower side - and turned "off" everywhere else.
"Without ta-siRNAs, leaves look like needles, because they lack an upper side," Timmermans says. "But we didn't understand how they set up patterning." This raised the question, in other words, of why ta-siRNAs only switch off ARF3 on the upper side of leaves. The CSHL team's finding that these RNA molecules seem to act as morphogens now solves the puzzle.
"Establishment of a gradient of mobile small RNAs can create profound differences between neighboring cells by altering their gene activity patterns," Timmermans says. "This is a neat way of dividing a cluster of cells into distinct sections with sharply drawn boundaries."
The top-to-bottom, abundant-to-rare distribution, or "concentration gradient," of ta-siRNAs ensures that the activity of ARF3 is strongly inhibited in the leaf's top half, but mildly or hardly affected at the bottom, thus creating a sharp boundary between leaf sections with different fates.
Ta-siRNA biogenesis is spatially controlled
In addition to mobility, the team attributes the unique distribution pattern of these small RNAs to the way they are produced within the leaf - a biochemical process involving several complicated steps.
The small ta-siRNAs are generated from larger RNA strands called precursors that are snipped at specific sites. Two cellular ingredients ensure that the cuts occur in the right place: a microRNA molecule called miR390 that specifies the location of the first cut, and an enzyme called ARGONAUTE7 (AGO7) that ferries miR390 to this location and creates the cut.
The CSHL team found that although miR390 is present in all cells of the leaf, the precursors and ARGONAUTE7 are strictly restricted to only the cells in the two uppermost layers. The ta-siRNAs are therefore generated exclusively in these upper cell layers, from where they move to the lower side of the leaf, accumulating as a gradient.
Thus, besides identifying the first example of a morphogen-like small RNA signal, Timmermans and her team have also shown that the location of the various biochemical ingredients required for small RNA activity can impact pattern formation. Together, their discoveries explain how mobile small RNAs can generate patterns during development.
More information: "Pattern Formation via Small RNA Mobility" appears in the March 1st issue of Genes and Development. The full citation is: Daniel H. Chitwood, Fabio T. S. Nogueira1, Miya D. Howell, Taiowa A. Montgomery, James C. Carrington, and Marja C. P. Timmermans.
Source: Cold Spring Harbor Laboratory (news : web)