How useful are microsatellites?
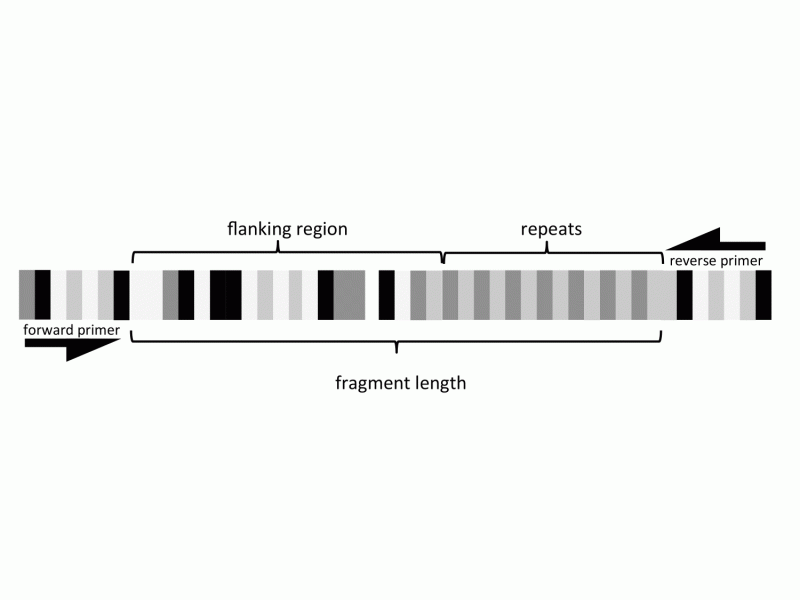
Understanding genetic diversity and evolutionary dynamics of organisms can have important implications for designing successful conservation efforts and understanding potential effects of future climate change. For scientists seeking to illuminate these dynamics and uncover genetic diversity within plant species, microsatellites are an often-used tool. These repeated segments of DNA sequences offer a glimpse into the evolutionary history of organisms at a very fine scale, providing a valuable data source for population-level studies.
"These markers aren't without potential problems, however," warns Dr. Charlotte Germain-Aubrey, a scientist in the Florida Museum of Natural History and lead author of a study in a recent issue of Applications in Plant Sciences. The study, a collaboration between scientists at the University of Florida, aims to provide a deeper understanding of the utility—and problems—of using fragment lengths, repeats, and flanking regions of microsatellite markers for conservation genetic studies.
"Several studies have examined and reported biases associated with the use of microsatellite genotyping," says Germain-Aubrey. "The widespread use of these markers for genetic studies led us to further explore the utility and problems associated with using microsatellites. We used an endangered plant species, Polygala lewtonii, as a test case."
The problem with microsatellites essentially boils down to whether or not you can confidently infer the relative number of repeats between individual plants. A problem arises when a mutation in the flanking region causes a change in the overall fragment length. When only considering fragment length, such a mutation can mimic the addition (or deletion) of a repeat, leading to erroneous conclusions. This is termed homoplasy—similarities in fragment length are not due to their shared ancestry but, rather, to an unseen change in an area adjacent to the repeats.
By sequencing the flanking regions, Germain-Aubrey and colleagues were able to quantify the amount of homoplasy in their data set. "Contrary to previous thinking, we found that homoplasy can profoundly affect studies working within even the most recently diverged populations," she explains.
An additional important result from this study is that researchers need to be wary when using microsatellite markers that were designed for a different species. The team found that the amount of homoplasy becomes increasingly problematic when considering multiple species or using markers designed for a taxon other than the target species. "Often, it's tempting to use markers that were previously developed for a closely related species. We strongly suggest that when doing this, researchers take the time to sequence several individuals from their study species using these markers before forging ahead with the study," concludes Germain-Aubrey.
More information: Charlotte C. Germain-Aubrey et al. Are Microsatellite Fragment Lengths Useful for Population-Level Studies? The Case of (Polygalaceae) , Applications in Plant Sciences (2016). DOI: 10.3732/apps.1500115
Journal information: Applications in Plant Sciences
Provided by Botanical Society of America