Team finds a new cellulose digestion mechanism by a fast-eating enzyme
Researchers at the Energy Department's National Renewable Energy Laboratory (NREL) have discovered that an enzyme from a microorganism first found in the Valley of Geysers on the Kamchatka Peninsula in Russia in 1990 can digest cellulose almost twice as fast as the current leading component cellulase enzyme on the market.
If the enzyme continues to perform well in larger tests, it could help drive down the price of making lignocellulosic fuels, from ethanol to other biofuels that can be dropped into existing infrastructure. A paper reporting this finding, "Revealing Nature's Cellulase Diversity: The Digestion Mechanism of Caldicellulosiruptor bescii CelA" appears in the journal Science.
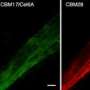
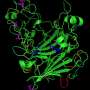
The bacterium first found in heated freshwater pools, Caldicellulosiruptor bescii, secretes the cellulase, CelA, which has the complex arrangement of two catalytic domains separated by linker peptides and cellulose binding modules.
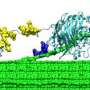
NREL researchers put CelA to the test and found that it produced more sugars than the most abundant cellulase in the leading commercial mixtures, Cel7A, when acting on Avicel, which is an industry standard to test cellulose degradation. They found that CelA not only can digest cellulose in the more common surface removal, but that it also creates cavities in the material, which leads to greater synergy with more conventional cellulases, resulting in higher sugar release.
The bacteria that secrete the promising CelA thrive in temperatures of 75 to 90 degrees Celsius (167-194 degrees Farenheit).
"Microorganisms and cellulases operating at such high temperatures have several biotechnological advantages," said NREL Scientist Yannick Bomble, one of the paper's authors.
"CelA is the most efficient single cellulase we've ever studied – by a large margin," Bomble said. "It is an amazingly complex enzyme, combining two catalytic domains with three binding modules. The fact that it has two complementary catalytic domains working in concert most likely makes it such a good cellulose degrader."
Most commercial operations use enzyme cocktails, a combination of 15 to 20 different enzymes, to turn plant material into the sugars that are valuable to the biofuels industry. In most such cocktails, one type of enzyme, Cel7A, does the largest amount of work.
When researchers compared CelA to Cel7A, they discovered that at its optimal temperature of 50 degrees Celsius (122 degrees F), Cel7A achieves only 50% of the performance of CelA when converting Avicel.
CelA was discovered 15 years ago, but until this recent work, all that was known about this complex protein was its general architecture and that it had the ability to degrade cellulose.
This organism, initially grown on biomass by scientists from the University of Georgia, was used to produce extracellular enzymes (enzymes that function outside of the cell). Those extracellular enzymes later were purified and characterized at NREL using techniques including performance assays, advanced imaging, X-ray crystallography, and modeling on supercomputers.
NREL scientists found that CelA is not only very active on cellulose, but also attacks xylose. That could mean that levels of enzymes that specialize in removing xylose in commercial cocktails could be lowered, translating to lower costs.
If an enzyme can produce sugars more efficiently, it means lower cost for the enzyme cocktail, which is a major cost driver in the process of converting biomass into fuel.
The findings have important implications for industry, but also were fascinating for the scientists. "We are learning a lot about the evolution of these cellulases, how they can thrive in extreme environments, and how they operate on biomass," NREL scientist and the paper's lead author, Roman Brunecky, said.
"This discovery could reshape the landscape of commercial cellulase cocktail design," said Paul Gilna, director of the BioEnergy Science Center, which provided the funding for this work. It is one of three Bioenergy Research Centers supported by the Office of Biological and Environmental Research in Energy Department's Office of Science.
More information: www.sciencemag.org/content/342/6165/1513
Journal information: Science
Provided by National Renewable Energy Laboratory