February 20, 2012 report
Oxford Nanopore announces groundbreaking GridION and MinION gene sequencers

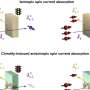
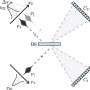
(PhysOrg.com) -- Oxford University spinoff company, Oxford Nonopore has announced at this year’s Advances in Genome Biology and Technology conference in Florida, two new machines for sequencing genes. Of particular note is the MinION, a machine small enough to fit in the hand which can be plugged into a laptop’s USB port. The other, the GridION, is a larger version that can be stacked to increase processing power. Both rely on a technology known as strand sequencing whereby a nanopore (engineered protein) is used to pull strands of DNA through a hole where a microchip measures minute changes in the electrical current in the membrane around it as individual bases, or pairs are pulled through. Because of the way it is done, much longer sections of DNA can be examined at once, doing away with the need to examine small sections independently and then knitting the results together with a computer afterwards.
Sequencing of genes is a process where the chemical order of DNA units (T, C, G and A) are determined. Doing so helps researchers and doctors determine inherited traits in plants and animals. It is an area of science that has been in the news of late as it is a hotbed of excitement for investors. This announcement by Oxford Nonopore comes as rather a shock to the established players in the field, American companies, Illumina and Life Technologies.
In addition to their small size, the new devices are able to perform sequencing faster than previous machines. Representatives of Oxford Nonopore say if 20 units (adding up to roughly $5000) are connected together the GridION can sequence an entire human genome in just fifteen minutes. In comparison, Life Technologies’ latest product, the Ion Proton Sequencer, at a price of almost $150,000, takes twenty four hours.
But it’s the MinION that is causing the most excitement in the scientific community. At just $900, any researcher anywhere could take a sample, in the field even, slip it into the device, then plug it into a laptop, and almost instantly have information about small genome samples. Seed research companies could use it to analyze crops in a field, for example, to see if they have mixed without outside sources, meat inspectors could use it test for different types of microorganisms, biologists could use it to look for small changes in genes over generations. The number of applications are literally too many to envision.
One dark spot in an otherwise rosy picture is the fact that the devices have a four percent error rate. Too high for many applications, though the company says it believes it can get the rate down significantly before the product is released sometime this year.
More information: Company's press release
© 2011 PhysOrg.com