Reversing the conventional DNA wisdom
(PhysOrg.com) -- The copying of DNA's master instructions into messenger molecules of RNA, a process known as DNA transcription, has always been thought to be a unidirectional process whereby a copying machine starts and moves in one direction. But in work that represents a fundamental shift in scientists' understanding of the phenomenon, MIT researchers have found evidence that two DNA copying machines frequently start from the same site and move in different directions.
MIT Institute Professor Phillip Sharp and his colleagues, who report the results in the Dec. 4 early online edition of Science, believe this new mechanism may play a role in keeping genes poised for transcription.
"People have been studying transcription for a long time and never seen this kind of transcription before," said Amy Seila, a postdoctoral associate in Sharp's lab and lead author of the paper.
DNA, which is housed within the nucleus of cells, controls cellular activity by coding for the production of enzymes and proteins. The genetic information in DNA is not directly converted into proteins, but must first be copied, i.e. transcribed, into RNA.
During normal transcription, an enzyme called RNA polymerase binds to a gene's transcription start site, and then proceeds downstream along the DNA, copying the sequence and producing messenger RNA that carries the gene's instructions.
In the new scenario, a second RNA polymerase is believed to bind near the starting point and move upstream, producing a short RNA sequence that doesn't code for any proteins.
"It looks like we have a polymerase that binds as we thought, but we also see a polymerase that appears to be pointing in the wrong direction, going upstream from the transcription start site," Seila said.
The researchers believe that upstream and downstream RNA polymerases, or divergent polymerases, are "paused" on the DNA and keeping the transcription start site open, so the gene can be easily accessed and transcribed.
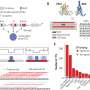
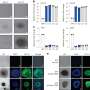
The MIT team postulated this divergent transcription after discovering a new class of small RNA, which they dubbed TSSa-RNA because it is associated with transcription start sites.
The researchers found from analysis of tens of millions of short RNA sequences that many of these RNAs were clustered just upstream of gene transcription start sites, suggesting that DNA transcription is occurring in the reverse direction. Many of the genes where these short RNAs were found are very active, supporting the theory that this mechanism helps promote gene transcription.
The researchers observed this phenomenon in several kinds of cells in both humans and mice, leading them to believe that it is universal.
A paper by another researcher, John Lis of Cornell University, offers independent evidence for the same theory. In a paper also appearing online in Science this week, Lis reports finding RNA polymerases sitting on genes both upstream and downstream from transcription start sites, waiting to be activated.
Sharp's lab plans to undertake further experiments to determine what roles these transcription start site-associated RNAs play in gene regulation, and whether they might interact with other types of small RNA such as microRNA.
"This is only the beginning of the story. It's certainly not the end of the story," said Sharp.
Other authors of the paper are J. Mauro Calabrese, a former graduate student in Sharp's lab; Stuart Levine and Peter Rahl, postdoctoral associates in the Whitehead Institute; Ryan Flynn, a junior biology major; Richard Young, professor of biology and member of the Whitehead Institute; and Gene Yeo of the University of California at San Diego.
Provided by MIT