Scientists build a better DNA molecule
Building faultless objects from faulty components may seem like alchemy. Yet scientists from the Weizmann Institute’s Computer Science and Applied Mathematics, and Biological Chemistry Departments have achieved just that, using a mathematical concept called recursion. 'We all use recursion, intuitively, to compose and comprehend sentences like ‘the dog that chases the cat that bit the mouse that ate the cheese that the man dropped is black,’' says Prof. Ehud Shapiro.
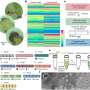
Recursion allows long DNA molecules to be composed hierarchically from smaller building blocks. But synthetic DNA building blocks have random errors within their sequence, as do the resulting molecules. Correcting these errors is necessary for the molecules to be useful. Even though the synthetic molecules are error prone, some of them are likely to have long stretches that do not contain any faults.
These stretches of faultless DNA can be identified, extracted, and reused in another round of recursive construction. Starting from longer and more accurate building blocks in this round increases the chances of producing a flawless long DNA molecule.
The team, led by doctoral students Gregory Linshiz and Tuval Ben-Yehezkel under the supervision of Shapiro, found in their experiments that two rounds of recursive construction were enough to produce a flawless target DNA molecule. If need be, however, the error correction procedure could be repeated until the desired molecule is formed.
The team’s research, recently published in the journal Molecular Systems Biology, provides a novel way to construct faultless DNA molecules with greater speed, precision, and ease of combining synthetic and natural DNA fragments than existing methods. 'Synthetic DNA molecules are widely needed in bio-logical and biomedical research, and we hope that their efficient and accurate construction using this recursive process will help to speed up progress in these fields,' says Shapiro.
Source: Weizmann Institute of Science