This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
proofread
New method for marking neurotransmitter receptors in living animal brains
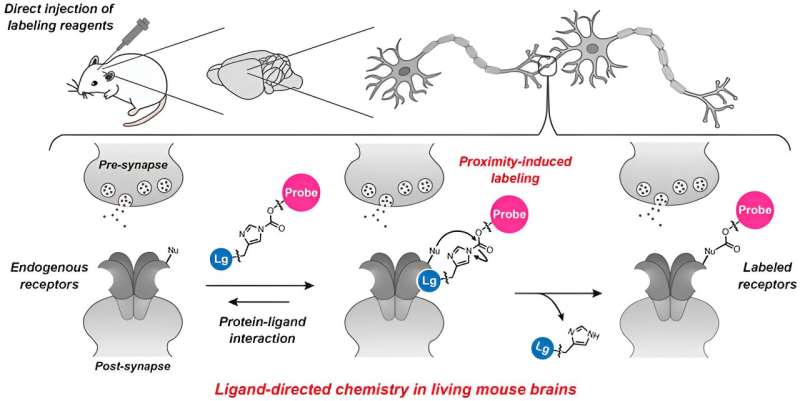
Researchers have developed a new method of labeling naïve neurotransmitter receptor proteins in living animal brains.
The work is published in the journal Proceedings of the National Academy of Sciences. The researchers include Kyoto University Professor Itaru Hamachi, Associate Professor Hiroshi Nonaka, Associate Professor Kiyoshi Sakamoto, and doctoral student Kazuki Shiraiwa
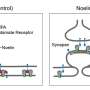
Conventionally, proteins emitting fluorescence (fluorescent proteins) were expressed in a state fused to proteins produced by genetic modification. However, due to concerns about (1) genetic modification being necessary, (2) functional hindrance of the fused proteins to the target protein, and (3) concerns about defects during expression, there was a demand to develop technology to mark (label) proteins in a more natural state.
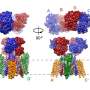
For the first time in the world, the research group succeeded in chemically labeling natural neurotransmitter receptors in mouse brains without genetic manipulation by using ligand-directed acylimidazole chemistry (LDAI chemistry). By pulse-chase analysis of AMPA-type glutamate receptors (AMPA receptors) in the postnatal developmental brain utilizing this method, it was determined that AMPA receptors that once performed a function moved to synapses that play different roles and are reused.
In principle, this technology can be applied not only to mice but also to other species. Developments can also be expected for many species, including primates such as marmosets, where labeling a target is difficult with genetic methodologies. Furthermore, it can be directly applied to model animal experimental systems (disease models, genetically modified mice, etc.) established to date by researchers around the world, and one can expect that relationships between pathology and receptor dynamics will be clarified.
In addition, this technology not only analyzes the fate (movement and lifespan) of proteins, but it can be useful for elucidating the function of natural proteins in individual animals by introducing various functional molecules in the future, and the research is ongoing.
This research was conducted in collaboration with Professor Shigeki Kiyonaka of Nagoya University, Professor Etsuo Susaki of Juntendo University, Professor Hiroki Ueda of the University of Tokyo, Professor Michisuke Yuzaki, Associate Professor Wataru Kakegawa, and Assistant Professor Itaru Arai of Keio University.
The ongoing research project aims to accurately elucidate information transmission and intercellular network formation within the nervous system and brain at the level of individual protein molecules by creating innovative chemical biology molecular techniques.
More information: Hiroshi Nonaka et al, Bioorthogonal chemical labeling of endogenous neurotransmitter receptors in living mouse brains, Proceedings of the National Academy of Sciences (2024). DOI: 10.1073/pnas.2313887121
Provided by Japan Science and Technology Agency (JST)