September 8, 2023 report
This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
trusted source
proofread
Downstream RNA hairpins found orchestrating mRNA translation
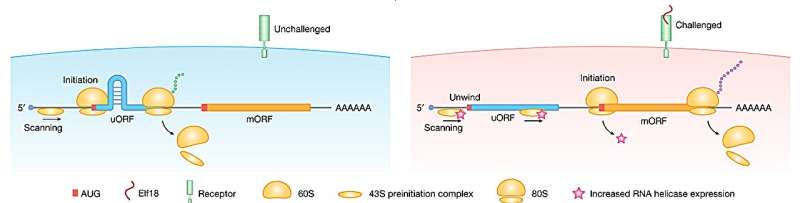
Research led by Duke University, Durham, has discovered a situation-dependent traffic jam in mRNA translation caused by RNA hairpins leading to higher translation of upstream start codons (uAUGs).
In a paper, "Pervasive downstream RNA hairpins dynamically dictate start-codon selection," published in Nature, the team describes their findings of a translational regulation mechanism in the plant Arabidopsis and consequent discovery of a similar mechanism in humans. A News & Views piece in the same journal issue discussed the work done by the team.
Any genome is constructed with instructions that inform when to start and stop the transcription of RNA segments. These segments can contain multiple start codons, which can then be selectively used as beginning points for the ribosome to start translating them into proteins. The current study has identified a mechanism by which the selection process between potential starting points takes place.
Ribosomal subunits assemble on an mRNA molecule and scan it for the AUG start codon to initiate protein synthesis. Sometimes, the first start codon encountered is used as initiation, and sometimes, it is not recognized. If it is not recognized, the scanning continues down the mRNA, where it may find an alternative starting point or a more easily recognized starting point (the main AUG).
The study discovered that if the mRNA has a hairpin loop located between two starting codons, the scanning process will be slowed down, providing more time for recognition of the upstream start codon (uAUG), leading to a higher translation rate from this starting point.
Arabidopsis seedlings were studied under an immune response triggered by the presence of elf18 (a fragment of a bacterial protein) to identify changes in codon selection. It found that ribosome activity at the initial upstream start codons decreased in response to elf18, suggesting that translation preferentially begins at the more easily recognized (main AUG) starting points under these conditions.
The study identified RNA helicases, specifically RH37-like helicases, as enzymes involved in unwinding hairpin structures near uAUGs during the immune response in Arabidopsis. Elevated levels of these helicases under immune response eliminated the traffic jam caused by the hairpin, which then increased translation from the alternative start codon and thus produced a different protein.
The "if this, then that" mechanism functions much like a logic gate, allowing the same mRNA segment to produce an alternative protein when the situation dictates.
The researchers introduced a hairpin structure downstream of a start codon in ATF4, a well-known mammalian stress-responsive gene with higher expression levels under cellular stress conditions. This condition inhibited the translation of ATF4 through enhanced translation initiation of a less recognized start codon.
The researchers performed in vivo analysis on a mutant version of the BRCA1 mRNA associated with breast cancer. The mutant BRCA1 mRNA is known to be regulated by having multiple start codons. The analysis detected significantly lower activities downstream of uAUG2 and uAUG3 using SHAPE-MaP analysis, indicating the presence of hairpin structures in mammalian transcripts.
The experimental results demonstrate that upstream start codons and downstream hairpin structure-mediated translation initiation mechanisms are not limited to plants but are also present and functional in human cells. This suggests a universal logic gate mechanism for start-codon selection in translation initiation across different organisms with both conserved evolutionary insights and future therapeutic targeting implications.
More information: Yezi Xiang et al, Pervasive downstream RNA hairpins dynamically dictate start-codon selection, Nature (2023). DOI: 10.1038/s41586-023-06500-y
Yizhu Lin et al, Dynamic regulation of messenger RNA structure controls translation, Nature (2023). DOI: 10.1038/d41586-023-02673-8
Journal information: Nature
© 2023 Science X Network