Study unravels the role of DNA methylation in oceanic microbial communities
DNA methylation is a biological process through which methyl groups are added to the DNA (genetic material). It is used as an epigenetic, i.e., a non-genetic strategy by prokaryotes to perform an array of functions such as gene regulation, repair, and protection against viral invasion using restriction-modification (RM) systems, which function as prokaryotic immune systems.
Until recently, studies related to DNA methylation have been restricted to microorganisms that can be cultured in laboratory settings. This has led to a poor understanding of its role in microbial ecology. It is, therefore, essential to conduct genome-wide epigenetic studies of environmental microbes, particularly those which cannot be cultured in the laboratory, but only thrive under natural conditions.
To this end, a team of researchers led by Professor Woo Jun Sul from Chung-Ang University and Dr. Hoon Je Seong (currently from Macrogen Inc.), South Korea has explored the differences in DNA methylation patterns across different members of the ocean microbial communities in the northwest Pacific Ocean.
Their study was published in Microbiome.
"[The] in-depth DNA methylation project began only in 2014, with the release of long-read sequencers. This sparked our curiosity and we wanted to apply it to microbial ecology. Hence, we used a metagenomics approach to explore DNA methylation in a community rather than at an organism level," says Prof. Sul while discussing the motivation behind their study.
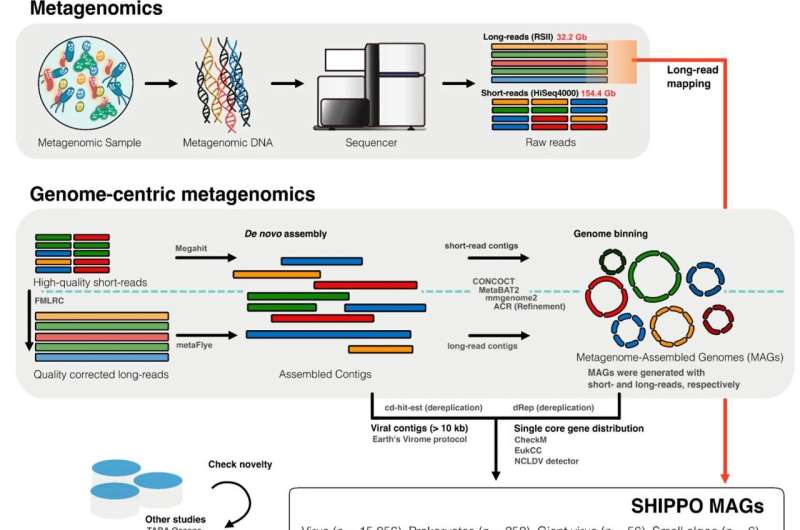
The hustle began back in 2015, when the large-scale Shipborne Pole-to-Pole Observations (SHIPPO) project was initiated by the Korea Polar Research Institute. It involved filtering out microorganisms from ocean surface samples across 10 different stations from the Pacific Northwest to the Bering Sea.
The team extracted DNA from these captured specimens and used short- and long-read sequencers to perform metagenomic sequencing. These sequences were then aligned using computational analysis to generate massive 15,056 viral (v), 252 prokaryotic (pro), 56 giant viral (gv), and 6 eukaryotic (eu) metagenome-assembled genomes (MAGs).
Upon further analyses, the team was surprised to find that nearly 95% of the sequenced proMAGs belonged to new taxa that could not be classified using existing genomic databases. "This finding clearly demonstrates the amount of potential this technique has, and how it could provide new insights into the genomes of unculturable ocean microbes," Prof. Sul explains.
Next, the team used this approach to explore the diversity of DNA methyltransferase (MTase) enzyme classes expressed by the genomes identified in the SHIPPO database.
They found that MTase II was the most common class of MTase expressed in these organisms. Interestingly, most of the proMAGs lacked complete RM systems due to the absence of restriction enzymes. Furthermore, the identification of methylated motifs across the ocean microbiome revealed unique DNA methylation patterns, which eventually led to the discovery of a distinct methylation profile in Alphaproteobacteria.
Next, the team used single molecule real-time (SMRT) sequencing to observe methylation patterns in Pelagibacter. They discovered heterogeneity in the methylation profile of the bacteria even at the "strain-level." This implies that dynamic cellular events occur within Pelagibacter in the surface waters of the northwest Pacific Ocean.
A comparative analysis of the bacterial and viral genomes also provided clues to their evolutionary patterns and interactions. The team found the presence of uneven methylation patterns in the Cand. P. Giovannoni NP1 genome, suggesting potential defense mechanisms used by this bacterium.
These findings have already paved the way for a new era of meta-epigenomics, which directly measures methylation in environmental microbes. The potential of studying the epigenome of various organisms at once is far-reaching.
Prof. Sul says, "Along with studies to identify methylation patterns of strains showing actual pathogenicity, our study also helps discover candidate targets to prevent pathogenicity in the environment. This can be of immense importance to the global public health systems by detecting pathogenic signals that threaten human health."
More information: Hoon Je Seong et al, Marine DNA methylation patterns are associated with microbial community composition and inform virus-host dynamics, Microbiome (2022). DOI: 10.1186/s40168-022-01340-w
Provided by Chung Ang University



















