Scientists provide structural insights into NaV1.7 modulation by inhibitors, to block pain signals to the brain
Chronic pain is an extremely common condition that affects about 20% of the general population. Given the shortage of effective and non-addictive analgesics, new anti-pain drugs are eagerly awaited. Voltage-gated sodium channel NaV1.7 plays an essential role in the transmission of pain signals to the brain, and multiple mutations in NaV1.7 have been directly linked to a variety of human pain disorders.
The blockade of NaV1.7 can inhibit pain sensation; thus, it represents an attractive target for potential non-addictive analgesics. However, NaV1.7 is a very challenging target for developing selective candidate drugs, partially owing to the high sequence similarity within the nine NaV channel isoforms.
Understanding the structural discriminations among NaV channel isoforms and the mechanism of how these inhibitors regulate NaV1.7 functions can support NaV1.7-related drug development. Zhang Jiangtao in Prof. Jiang Daohua's group from the Institute of Physics of the Chinese Academy of Sciences has reported on the cryo-EM structures of NaV1.7 in complex with three pore blockers, providing mechanistic insights into NaV1.7 modulation by pore blockers.
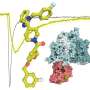
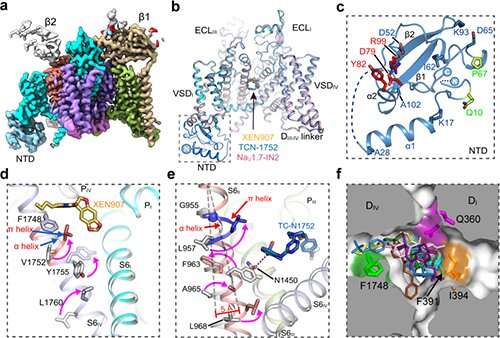
The researchers solved high-resolution cryo-EM structures of NaV1.7 complexed with three chemically distinct small molecule inhibitors XEN907, TCN-1752, and NaV1.7-IN2, respectively.
The structure revealed the previously unresolved N-terminus domain (NTD) of NaV1.7, explaining that the conserved NTD is critical for NaV channel function.
In addition, the structures confirmed that the central cavity of NaV1.7 accommodates multiple drug binding sites, which can directly block the channel. Two of the three inhibitors also caused local conformational rearrangements of the S6 helix, that further affect the channel function.
The XEN907 caused a α-helix to the π-helix transition in S6IV of NaV1.7, which significantly slowed down the recovery of NaV1.7 from fast inactivation. The binding of TC-N1752 indirectly caused a local shift from the α-helix to the π-helix in S6II of NaV1.7, and shifted the S6II helix approximately 5 Å toward the activation gate, leading to a completely closed activation gate, and stabilizing NaV1.7 in the inactivated state.
Further structural analysis revealed that the inhibitor binding sites located in the central cavity are highly conserved among the nine NaV channel isoforms. Therefore, it is very challenging to achieve subtype-selective drugs binding in the central cavity of NaV channels.
This study suggests that future efforts on searching for NaV1.7-selective inhibitors should focus on the regions that are relatively less conserved, such as the voltage-sensing domain.
This study, titled "Structural basis for NaV1.7 inhibition by pore blockers," was published in Nature Structural & Molecular Biology.
More information: Jiangtao Zhang et al, Structural basis for NaV1.7 inhibition by pore blockers, Nature Structural & Molecular Biology (2022). DOI: 10.1038/s41594-022-00860-1
english.cas.cn/newsroom/resear … 1127544501987768.pdf
Journal information: Nature Structural & Molecular Biology
Provided by Chinese Academy of Sciences