What survives, thrives and dominates over a thousand generations? The answer might be even more complex than thought

Aa team of scientists, led by Harvard researchers, has used a new method of DNA "re-barcoding" to track rapid evolution in yeast. The new approach, published in Nature, advances the field of organismic and evolutionary biology and holds promise for real-world results.
The potential impact of the work can be illustrated using the example of flu vaccines. An accurate prediction of what strains of influenza will dominate over the next year is necessary to ensure the vaccines produced are useful. Such prediction relies on tracking evolution.
"We have the sequence of all these flu strains, and we're watching their evolution. What you should be able to do is look at how they've evolved in the past and be able to predict into the future what is going to win and what is going to lose. The problem is, we don't know how to do that prediction," explained Michael Desai, Professor of Organismic and Evolutionary Biology (OEB) and of Physics at Harvard.
Desai, in whose lab the study was conducted, said that the questions are basic: "There is this swarm of mutations that are constantly happening," he said. "How do they battle it out, and what determines who wins?"
"We have been taught that evolution 'is slow' and involves the 'survival of the fittest,' added Alex N. Nguyen Ba, a post-doctoral fellow in Desai's lab. "It turns out that molecular evolution doesn't work that way. It's actually much faster than how we've been taught. This makes evolution way more complex than what has been anticipated." Nguyen Ba is one of three co-lead authors of the new study, along with Ivana Cvijović and José I. Rojas Echenique
Such evolution has been posited mathematically over the past two decades. However, previous lab experiments have not been able to prove or disprove the theory. Rather, they have only been able to examine the process with high resolution over a short period of time, or with low resolution over a long period of time. Collectively, Desai explained, the paper's authors—who include Katherine R. Lawrence of MIT and Harvard's Artur Rego-Costa, along with Xianan Liu of Stanford and Sasha F. Levy of SLAC National Accelerator Laboratory - have done both other kinds of studies.
This new study does both.
"We can identify every single relevant beneficial mutation," said Nguyen Ba, citing new technology that allowed the research team to follow specific genomes (or lineages) for approximately a thousand generations.
Cvijović, formerly a graduate student in Desai's lab and now a researcher at Princeton, said the research could have gone on indefinitely: "A thousand generations is about three months of growth in our conditions. That's enough time to see big changes happening."
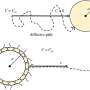
Such in-depth, long-term research was possible because of a technological advance in the methodology that allowed what Nguyen Ba called the "re-barcoding" of DNA.
Using an enzyme to place a marker, the "barcode," at a specific DNA site, the researchers were able to follow the DNA of yeast through multiple generations. By re-tagging and re-barcoding subsequent generations to record their lineage, the team could then observe how this DNA was transmitted, noting what survived, and what thrived—or came to dominate—as generations passed.
What they discovered included a few surprises.
According to the existing theory, the "fittest" DNA would be that which showed up most frequently in subsequent generations. However, the scientists observed "fluctuations" that the theories could not account for.
"Mutations and genotypes that seem to have fallen behind can leapfrog and dominate," said Cvijović.
What that means, she says, will be the subject of future research. However, it implies that evolution is, indeed, even more complex than previously thought.
"Our experiment suggests there may be a wide range of a large number of strongly beneficial mutations," she said. "And their benefits are both very strong and very different from one another."
More information: High-resolution lineage tracking reveals travelling wave of adaptation in laboratory yeast, DOI: 10.1038/s41586-019-1749-3 , nature.com/articles/s41586-019-1749-3
Journal information: Nature
Provided by Harvard University