August 8, 2019 feature
Ex2SM: A text mining method to detect repeated strings in biological sequences
For several years, researchers have been trying to use computational methods for exact string matching, which entails identifying repeating patterns in long strings of text or digits. This is because tools that can automatically identify these repeating patterns could have numerous important applications in fields such as genetics and biology.
Konstantinos F. Xylogiannopoulos, an associate researcher at the University of Calgary, has recently developed a new text mining technique that can detect every possible repeated string in multivariate biological sequences. His work could help experts in their search for advanced treatments for serious diseases, including genetics-related ones, such as cancer or Alzheimer's.
"My research was inspired by a paper about pattern matching in DNA sequences," Xylogiannopoulos said. "Although I had general knowledge in biology, I had never considered the complexity of the problem from a computer science perspective, because of the size of a DNA sequence. Since then, I devoted myself to trying to simplify pattern detection for big data by optimizing my algorithm for detecting repeated patterns."
Shortly after he started researching string matching in DNA sequences, Xylogiannopoulos made an interesting observation. He found that many well-known computer science problems can be efficiently solved by transforming them into a repeated pattern detection problem, irrespective of their size or complexity.
"For example, a few months ago, Google announced the calculation of the first 31 Trillion digits of pi." Xylogiannopoulos said. "Yet, since 2016, I have detected the two longest repeated patterns that exist in the first 1 trillion digits of pi, something practically impossible with other algorithms. Thanks to Google pi-api it is very easy now to verify my findings."
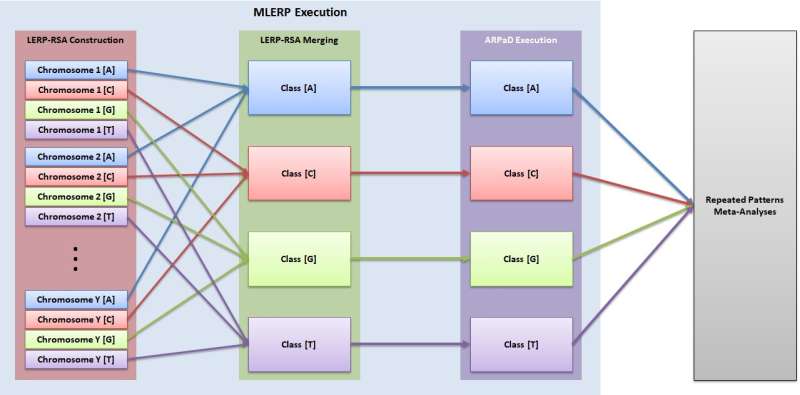
In his recent study, Xylogiannopoulos developed a pipeline of execution of advanced data structures and algorithms that can be used for text mining. This technique, called Ex2SM, achieves string matching via a series of important steps.
"Firstly, the technique creates the longest expected repeated pattern reduced suffix array (LERP-RSA) for suffixes of a predefined length (LERP) using several attributes of the LERP-RSA, such as classification based on the DNA alphabet (A, C, G and T)," Xylogiannopoulos explained. "Then, the all repeated pattern detection (ARPaD) algorithm is executed to detect every pattern that exists at least twice. For patterns that are found to have a length of exactly LERP, the Moving LERP algorithm is executed to create a new LERP-RSA and execute ARPaD. The process is repeated, in parallel, until all patterns, regardless of length, have been discovered."
The method developed by Xylogiannopoulos allows for classification and parallelization. Moreover, it can be executed on completely isolated and different hardware, software or cloud systems. In other words, Ex2SM works well irrespective of hardware limitations or dataset sizes. What truly differentiates it from other existing methods, however, is that it is input agnostic, i.e. it does not require input strings in order to search for them.
"Let's imagine that we have a book and each time we need to search for a word or phrase in a chapter we need to repeat the searching process using any pattern matching algorithm," Xylogiannopoulos said. "In this context, Ex2SM could be seen as a process that generates an 'index' of all repeated words, phrases etc. in the chapter by executing the process just once. This index allows us to achieve fast outputs for any kind of meta-analyses by executing complex and targeted queries directly on the results (usually a simple binary search), something that, to the best of my knowledge, no other methodology or algorithm has achieved so far."
Interestingly, Ex2SM can be applied to a variety of different tasks. For instance, it could be scaled up to analyze a simple system of chapters (e.g. a book), a complex multivariable system (e.g. a library or collection of books), and even a high-multidimensional system (e.g. a universal library or collection of libraries). When applied to the field of bioinformatics, on the other hand, the new technique could be used for string matching of chromosomes, human genomes, a collection of human genomes, and ultimately even genomes for all species within a universal database.
In his study, Xylogiannopoulos specifically wanted to highlight the potential Ex2SM for deep pattern detection and data mining within the field of bioinformatics. Remarkably, he successfully used his method to analyze the entire human genome and unveil repeating patterns. This allowed him to observe, for example, that there is a specific type of long repeated patterns that exists only in certain chromosomes.
"More complicated studies could also be performed using Ex2SM," Xylogiannopoulos said. "For example, we know that there is a connection between breast and prostate cancer, two entirely different organs, because of the BRCA1 and BRCA2 genes. What this method can disclose is a possible interconnectivity among different diseases because of common 'sections' in DNA, RNA, protein sequences etc. that could probably connect more diseases and their treatments."
Using Ex2SM, Xylogiannopoulos was able to detect all repeated strings in the human genome with a length of up to 50 characters. This would be virtually impossible to achieve using most existing algorithms and text mining techniques, due to the vast amount of possible permutations. For instance, to find all 50-character-long patterns, a brute force algorithm would need to process millions of trillions of permutations separately.
In the future, Ex2SM could help experts in bioinformatics and biology to broaden current understanding of genomics behavior and unveil important genetic links between diseases. Xylogiannopoulos is currently using his technique to analyze human genes and detect all patterns repeated in them. His studies have already produced a vast pool of results, which are so extensive that he is unable to share all of them with the public.
"I am processing an idea to create a platform, possibly in cooperation with an institution, that will give on line access to the results for researchers and domain experts," Xylogiannopoulos said. "I am also working on transforming my methodology to cooperate with deep learning neural networks for applications in image analysis, recommendation systems, natural language processing etc."
The different algorithms and components that power Ex2SM (i.e. LERP-RSA and ARPaD) have already been used to develop solutions for problems rooted in a variety of different fields, including text mining, web/network analytics and security, data streaming, time series analysis, clustering, classification, and many more. Xylogiannopoulos is also collaborating with colleagues at other universities to expand and improve these solutions, while also developing new ones.
More information: Exhaustive exact string matching: the analysis of the full human genome. arXiv:1907.11232 [cs.DS]. arxiv.org/abs/1907.11232
© 2019 Science X Network