Addressing South Africa's cancer reporting delay with machine learning

Cancer registries hold vital data sets, kept tightly encrypted, containing demographic information, medical history, diagnostics and therapy. Oncologists and health officials access the data to understand the diagnosed cancer cases and incidence rates nationally. The ultimate goal is to use this data to inform public health planning and intervention programs. While real time updates are not practical, multi-year delays make it challenging for officials to understand the impact of cancer in the country and allocate resources accordingly.
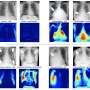
Unstructured pathology reports contain tumor specific data and are the main source of information collected by cancer registries. Human experts label the pathology reports using International Classification of Disease for Oncology (ICD-O) codes spanning 42 different cancer types. The combination of manual processes and the magnitude of reports received annually leads to a four-year lag for the country. In comparison, there is nearly a two-year delay in the United States.
In 2016, when we inaugurated our new IBM Research lab in Johannesburg, we took on this challenge and are reporting our first promising results at Health Day at the KDD Data Science Conference in London this month.
Our goal from the beginning was to apply deep learning to automate cancer pathology report labeling to speed up the reporting process. Working with the National Cancer Registry in South Africa, we used 2,201 de-identified, free text pathology reports and I am proud to report that our paper demonstrates 74 percent accuracy – an improvement over current benchmark models. We believe we can get to 95 percent accuracy with more data.
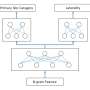
We employed hierarchical classification with convolutional neural networks, although this was not our first choice. We initially started exploring multiclass and binary convolutional neural networks models, but the results were not promising and I nearly quit in frustration. Eventually, with the advice and support of my colleagues, we cleaned up the text, refined the feature engineering process and improved it to 60 percent. This result was an improvement, but we knew we needed 90-95 percent to make it trustworthy enough for the real world.
After more research and exploration, we thought about reducing the complexity of the multiclass problem, which led us to create a state-of-the-art hierarchical deep learning classification method based on the hierarchical structure of the oncology ICD-O coding system. Thus, we used a combined approach to identify class hierarchy and validate it using expert knowledge to achieve better performance than a flat multiclass model for classification of free text pathology reports.
Our work is of course not done yet; we need to reach above 95 percent accuracy, and we think this is possible with more data, which will be provided by our partners at the National Cancer Registry. Once we get this, we think South Africa can be the best in the world in terms of cancer reporting, which is significant particularly because it's been reported that my country will see a 78 percent increase in cancer by 2030.
More information: Hierarchical Deep Learning Ensemble to Automate the Classification of Breast Cancer Pathology Reports by ICD-O Topography, Waheeda Saib, David Moinina Sengeh, Gciniwe Dlamini and Elvira Singh
Provided by IBM
This story is republished courtesy of IBM Research. Read the original story here.