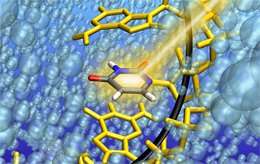
RNA strand enables trapping of uracil in the critical state
Researchers led by Regina de Vivie-Riedle, a professor of theoretical chemistry at LMU Munich, have found indications for a base-independent mechanism that can decrease the photostability of the RNA base uracil.
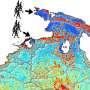
The building blocks of life, namely the five nucleobases adenine, guanine, cytosine, thymine and uracil that make up the genetic code, are susceptible to damage by UV radiation. After photoexcitation, they can undergo chemical reactions with their neighbors in a DNA or RNA strand, causing dangerous mutations that eventually increase the risk for skin cancer.
Fortunately, all five nucleobases have ways of rapidly dissipating the energy deposited in the critical excited state. This relaxation process happens on a femtosecond timescale, faster than competing chemical reactions can occur, thereby preventing photodamage in most cases. Obstruction of these ultrafast pathways increases the chance that harmful photoproducts will be formed, since the nucleobase remains in the excited state for longer.
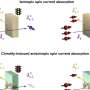
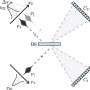
Up until now, such delayed relaxation into the ground state has mostly been attributed to the delocalization of excited states across several nucleobases. Professor Regina de Vivie-Riedle with her research team at LMU have now found indications for another mechanism that can occur on a single nucleobase, without the need for excited-state delocalization. Using state-of-the-art quantum dynamical methods that take the complex RNA environment into account, they found that the sterical influence of the RNA strand can obstruct the molecular motion necessary for the ultrafast relaxation of uracil and trap the nucleobase in the excited state for several picoseconds – long enough for harmful chemical reactions to occur. The work appears in the Journal of the American Chemical Society.
By considering different base sequences in their simulations, they also investigated whether specific combinations of neighboring nucleobases differ in their effects on the photostability of uracil. The results indicate that the described mechanism is a rather general effect of the molecular RNA environment, and occurs independently of any particular base sequence.
The use of computer simulations allows one to isolate the effects of different factors, some of which may be inaccessible to experimental investigation. Theoretical models therefore enable a more complete understanding of nature and are an integral part of modern chemistry. The computational effort for this study was considerable, and the authors gratefully acknowledge the resources provided by the Leibniz Supercomputing Center in Garching.
More information: Sebastian Reiter et al. RNA Environment Is Responsible for Decreased Photostability of Uracil, Journal of the American Chemical Society (2018). DOI: 10.1021/jacs.8b02962
Provided by Ludwig Maximilian University of Munich