Soil bacterium tapped for penicillin guard duty
A common soil bacterium may hold the key to preserving the germ-killing power of penicillin. U.S. Department of Agriculture (USDA) scientists in Peoria, Illinois, helped mass produce the antibiotic during World War II to treat Allied soldiers and later civilians. But decades of widespread use has since enabled some germs to develop resistance to it.
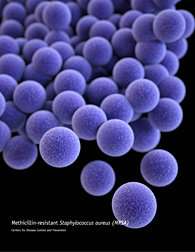
One strategy to counter this resistance being investigated by an Agricultural Research Service (ARS)-led team of scientists comes from soil-dwelling members of the bacterial group Streptomyces. These bacteria secrete a compound called "tunicamycin" to keep rival bacteria from reaching choice resources, like rotting plant material. This includes dispatching germs like the bacterium Staphylococcus aureus, which causes infection in humans and animals.
Tunicamycin works by forming holes in the cell walls of encroaching bacteria, causing them to burst open and die.
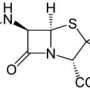
Researchers have known of tunicamycin for decades and were initially excited by its medical and veterinary prospects—especially as a way to overcome the resistance of some germs to penicillin-based drugs like oxacillin and methicillin. Problem was, tunicamycin also blocked a key protein in human and animal cells, undercutting its potential use in the ground war on germs.
Now, however, tunicamycin could get a new lease on life. ARS scientists have devised a method to retool the compound so that it poses little to no danger to human or animal cells but can still kills germs.
In laboratory trials, mixing the modified tunicamycin with oxacillin and other penicillin-based drugs made them 32 to 64 times more potent, according to ARS chemist Neil Price and colleagues at the agency's National Center for Agricultural Utilization Research (NCAUR) in Peoria, Illinois; the University of Illinois College of Medicine, and the Chinese Academy of Sciences.
Additionally, the modified tunicamycin didn't harm cultures of human and hamster cells when it was added to them in tests for toxicity, the team reported in a recent issue of the Journal of Antibiotics.
Price notes they used tunicamycin-producing Streptomyces bacteria taken from the same repository where the first mass-produced strain of the Penicillium mold is still kept. This repository is the ARS Microbial Culture Collection at NCAUR-designated in 2001 as an International Historic Chemical Landmark for its penicillin contributions.
More information: Neil PJ Price et al. Modified tunicamycins with reduced eukaryotic toxicity that enhance the antibacterial activity of β-lactams, The Journal of Antibiotics (2017). DOI: 10.1038/ja.2017.101
Provided by Agricultural Research Service