March 16, 2018 report
New device for studying cell lineage over multiple generations offers a way to measure effects of mutations
A team of researchers with members from several institutions in France has found a new way to study cell lineage over multiple generations. They developed a device (which they call a "mother machine") that is capable of separating out individual bacteria cells and watching as they divide over time and sometimes mutate. In their paper published in the journal Science, the group describes using the device to measure the percentage of E. coli bacteria that developed with lethal mutations.
As the researchers note, mutations in cells drive evolution, but they are also behind a host of problems that can harm humans. As part of the ongoing process of understanding mutations, it is important to know the rates at which they occur. Unfortunately, until now, a means for measuring such rates has not been available to researchers. In this new effort, the researchers developed a device to measure mutation rates in bacterial cells.
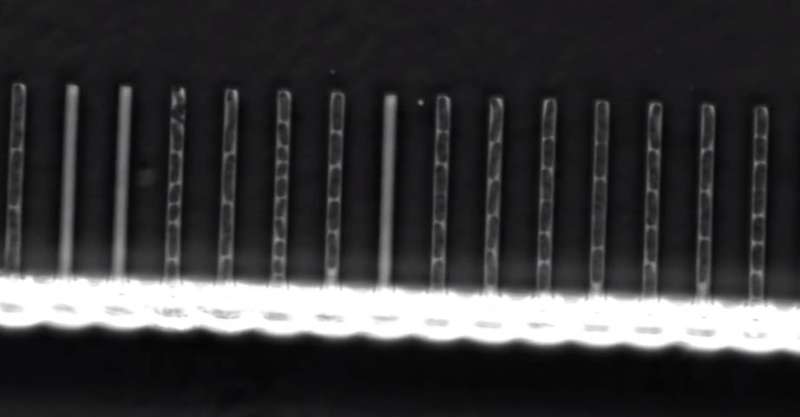
The device consists of a tube with multiple smaller tubes extending from one side, similar in appearance to a hair comb. Fluid moves along the central channel at the bottom and up into the teeth. Bacteria added to the fluid flow along with it, some of which are drawn up into the teeth. The teeth were designed to be small enough to allow passage of just one bacterium at a time. Once one of them moved up a single tube, it became trapped, allowing the researchers to watch it under a microscope. Those that became trapped were dubbed mother cells—when they divided, those that were released were called daughter cells—they were forced back into the main channel. Also, the bacteria introduced into the device had been engineered to become fluorescent when mutations occurred. The researchers then pumped fluid laden with bacteria through the system for a period of time, and the whole process was filmed with a time-lapse camera—it allowed for speeding up the action and for counting lethal mutations.
The researchers were able to watch approximately 200 generations of bacteria come and go—and in so doing, calculated that approximately 1 percent of the mutations that occurred were lethal.
More information: Lydia Robert et al. Mutation dynamics and fitness effects followed in single cells, Science (2018). DOI: 10.1126/science.aan0797
Abstract
Mutations have been investigated for more than a century but remain difficult to observe directly in single cells, which limits the characterization of their dynamics and fitness effects. By combining microfluidics, time-lapse imaging, and a fluorescent tag of the mismatch repair system in Escherichia coli, we visualized the emergence of mutations in single cells, revealing Poissonian dynamics. Concomitantly, we tracked the growth and life span of single cells, accumulating ~20,000 mutations genome-wide over hundreds of generations. This analysis revealed that 1% of mutations were lethal; nonlethal mutations displayed a heavy-tailed distribution of fitness effects and were dominated by quasi-neutral mutations with an average cost of 0.3%. Our approach has enabled the investigation of single-cell individuality in mutation rate, mutation fitness costs, and mutation interactions.
Journal information: Science
© 2018 Phys.org