July 27, 2017 report
Chemists deduce the correct structure of the A and B baulamycins
(Phys.org)—A team of chemists at the University of Bristol has correctly deduced the correct structure of the A and B baulamycins. In their paper published in the journal Nature, the team describes how they discovered that the baulamycins had been incorrectly structured and how they developed the new method for deducing the structure of flexible compounds in general. Severin Thompson and Thomas Hoye with the University of Minnesota offer a News & Views piece on the work done by the group in the same issue and explain how the new method could be used to help medical scientists identify natural compounds that might offer medical benefits.
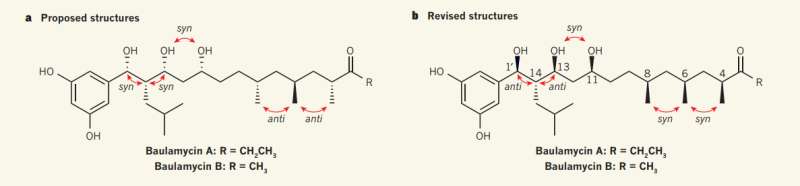
Despite their seemingly simple nature, molecules do not always display their true structure—they actually show a number of unique conformations. This makes it difficult for chemists attempting to understand or recreate them. Especially difficult is getting a handle on the true structure of flexible compounds. In this new effort, the researchers describe their discovery that the structure of A and B baulamycins had been incorrectly described by previous researchers and their efforts to find its correct structure, which led them to develop a new procedure to deduce the structure of a wide variety of natural compounds.
A and B baulamycins are polyketide antibiotics with long, flexible carbon chains. Because of their benefits, researchers have tried to create synthetic versions of them, but until now, have failed. The reason appears to be earlier incorrect descriptions of their structure. After making that discovery, the researchers set about determining the true structure.
The new method developed by the team involves using both computational methods and nuclear magnetic resonance spectra of the original material to figure out which configurations the compounds naturally take. They then made mixtures of possible isomers of baulamycin A with unequal ratios to identify which of the compounds matched the compound in its natural state. Once they had the correct structure, the team synthesized the compound, which can be used as an antibiotic. The team suggests the same process could be used by other researchers to deduce the correct structure of other natural complex molecules.
More information: Synergy of synthesis, computation and NMR reveals correct baulamycin structures, Nature (2017). nature.com/articles/doi:10.1038/nature23265
Abstract
Small-molecule, biologically active natural products continue to be our most rewarding source of, and inspiration for, new medicines. Sometimes we happen upon such molecules in minute quantities in unique, difficult-to-reach, and often fleeting environments, perhaps never to be discovered again. In these cases, determining the structure of a molecule—including assigning its relative and absolute configurations—is paramount, enabling one to understand its biological activity. Molecules that comprise stereochemically complex acyclic and conformationally flexible carbon chains make such a task extremely challenging . The baulamycins (A and B) serve as a contemporary example. Isolated in small quantities and shown to have promising antimicrobial activity, the structure of the conformationally flexible molecules was determined largely through J-based configurational analysis, but has been found to be incorrect. Our subsequent campaign to identify the true structures of the baulamycins has revealed a powerful method for the rapid structural elucidation of such molecules. Specifically, the prediction of nuclear magnetic resonance (NMR) parameters through density functional theory—combined with an efficient sequence of boron-based synthetic transformations, which allowed an encoded (labelled) mixture of natural-product diastereomers to be prepared—enabled us rapidly to pinpoint and synthesize the correct structures.
Journal information: Nature
© 2017 Phys.org