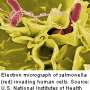
Team finds new antibiotic resistance gene in Salmonella from broiler chickens
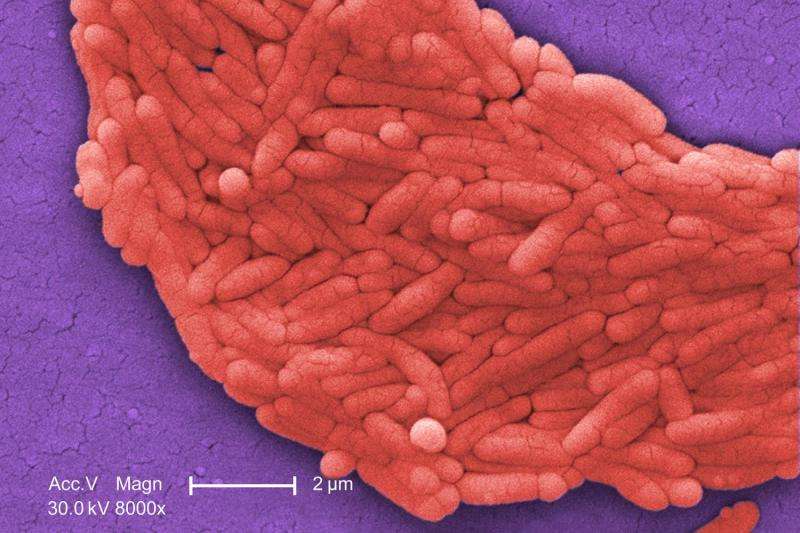
A team of investigators from Agriculture and Agri-Food Canada, Guelph, Ontario, has discovered a gene that confers resistance to the important broad-spectrum antibiotic, fosfomycin. The researchers found the gene in isolates of the pathogen, Salmonella enterica, from broiler chickens. The research is published in Antimicrobial Agents and Chemotherapy, a journal of the American Society for Microbiology.
The gene, dubbed fosA7, confers a high level of resistance to fosfomycin, which is otherwise a safe and effective agent for eliminating infections caused by multidrug resistant bacteria. (The "7" in fosA7 indicates that this is the seventh antibiotic resistant fosA gene that has been discovered.)
Currently, there is only limited fosfomycin resistance among Salmonella species, said corresponding author Moussa S. Diarra, PhD, Research Scientist in Food Safety at Agriculture and Agri-Food Canada. But the powerful resistance the fosA7 gene confers is worrisome, said Diarra. It could spread among different Salmonella serovars (a serovar is a strain of a species), as well as other bacterial pathogen species, via horizontal gene transfer, due to increased use of fosfomycin in both clinical and veterinary settings, said Diarra. Thus, "vigilant monitoring for the spread of fosfomycin resistance in bacteria, isolated from humans and animals, is needed."
With that in mind, the researchers tested the strength of the resistance the gene could confer on the closely related Salmonella enterica serovar Enteritidis. To do so, they cloned the gene, and inserted it into the chromosome of non-antibiotic resistant S. Enteriditis. Their worries were confirmed: the gene boosted the minimum concentration of fosfomycin required to inhibit reproduction in the microbe by more than 256-fold.
These results provided strong support for the hypothesis that fosA7 is, indeed, responsible for fosfomycin resistance, and that if fosA7 were transferred to plasmids—renegade pieces of DNA that can insert themselves into different bacteria—it could induce a high level of resistance in the recipient bacterial strain, according to the report.
The product of the fosA7 gene is an enzyme called glutathione-S-transferase. It inactivates fosfomycin by binding to it, and rupturing a molecular ring structure which is part of the antibiotic.
Fosfomycin resistance genes are often present in multidrug resistant bacteria. "This could further challenge the use of fosfomycin as an alternative treatment approach against urinary tract infections caused by both multidrug resistant E. coli, and blood infections from multidrug resistant Salmonella," said Diarra. So far, the investigators have found fosA7 only on S. enterica serovar Heidelberg and three other serovars of this species. A serovar is a strain of a species.
Salmonella enterica serovar Heidelberg—the strain in which the researchers found fosA7—is among the most common causes of human salmonellosis worldwide. The rise of resistance to multiple antibiotics, particularly to extended spectrum cephalosporins in Heidelberg has limited the number of therapeutic options against this Salmonella serovar. In the current study, the investigators found this resistance gene in all of 15 Salmonella Heidelberg isolates in their collection.
The American Society for Microbiology is the largest single life science society, composed of over 50,000 scientists and health professionals. ASM's mission is to promote and advance the microbial sciences.
ASM advances the microbial sciences through conferences, publications, certifications and educational opportunities. It enhances laboratory capacity around the globe through training and resources. It provides a network for scientists in academia, industry and clinical settings. Additionally, ASM promotes a deeper understanding of the microbial sciences to diverse audiences.
Journal information: Antimicrobial Agents and Chemotherapy
Provided by American Society for Microbiology