Trypanosomes evade detection by swapping coat proteins through chromosomal rearrangement
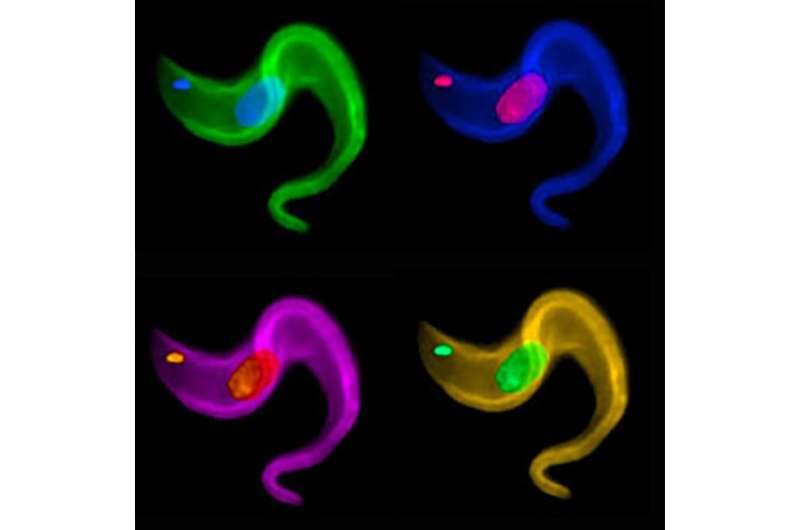
African trypanosomes establish deadly, chronic infections of trypanosomiasis in the bloodstream by using repetitive 70-bp regions in the genome to regularly change out the active coat protein gene. Galadriel Hovel-Miner and colleagues at Rockefeller University and George Washington University report this discovery and describe its role in the process that trypanosomes use to evade host antibodies on May 5, 2016 in PLOS Genetics.
Researchers have long known that a diverse group of pathogenic organisms can change their antigenic surface by changing the expression of their surface proteins. African trypanosomes, such as Trypanosoma brucei, are a model for this process whose genome contains more than 2,000 Variant Surface Glycoprotein (VSG) encoding genes. Only one VSG can be active at a time and a new VSG gene can be activated through chromosomal translocation into the actively expressed site. The current study asked the question, "how is a single gene selected from a repertoire of thousands?" The authors tested the prediction that a specific DNA repeat (termed 70-bp repeats) direct selection of the new VSG. By genetically manipulating the DNA repeats the authors were able to demonstrate that the 70-bp repeats promote selection of VSGs encoded throughout the genome and determine the essential DNA sequence required for this function.
The study provides a function for the 70-bp repeats and shows that they direct DNA pairing and recombination machinery away from the closest VSG genes and towards others in the archive. However, it remains unclear why one VSG is chosen over another.
If scientists could deactivate this process in natural populations, they could reduce the rates of chronic infections of African trypanosomiasis (sleeping sickness). The study may also provide insight into mechanisms of chromosome translocation in other species.
Dr. Hovel-Miner said of the publication, "We are excited that the findings in this manuscript finally provide a function for these intriguing DNA repeats that have been talked about since the earliest literature on antigenic variation. Furthermore, there is evidence here to suggest that the function of the 70-bp repeats the process of coat variation with the progression of cell cycle in a manner that give us exciting new insights in to the mechanistic basis of this critical pathogenic process."
More information: Hovel-Miner G, Mugnier MR, Goldwater B, Cross GAM, Papavasiliou FN (2016) A Conserved DNA Repeat Promotes Selection of a Diverse Repertoire of Trypanosoma brucei Surface Antigens from the Genomic Archive. PLoS Genet 12(5): e1005994. DOI: 10.1371/journal.pgen.1005994
Journal information: PLoS Genetics
Provided by Public Library of Science