May 19, 2016 report
A new way to create macrolides—from scratch—may help in battle against bacterial resistance
(Phys.org)—A team of researchers at Harvard University has found a way to create new macrolides—a class of drugs used to fight bacterial infections. In their paper published in the journal Nature, the team describes their approach and why they believe it might be useful in keeping ahead of bacterial resistance until something more revolutionary comes along. Ming Yan and Phil Baran with The Scripps Research Institute, offer a News & Views article outlining the work done by the team and why they believe the new technique may help medical researchers keep up with bacterial evolution.
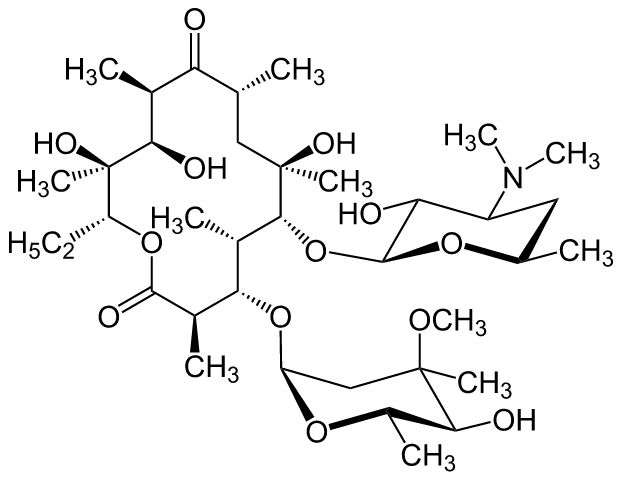
For many years, the go-to drugs of choice for treating a variety of bacterial infections has been macrolides, which are all drugs that have been created by altering a natural form of erythromycin—but in recent years, bacteria have become resistant to many of the drugs that have been developed. New ones have been slow in coming, due to the difficulty in finding new ways to alter the original bacteria which means R&D costs have been rising. This development has led to near-panic in the health community as it appears that if something does not change soon, the arsenal of weapons used to fight many infections will be vastly depleted. In this new effort, the researchers report that they have found a new way to create new variations of macrolides that does not require using native erythromycin as a source—instead, they create them from scratch. This development means that many more variants can be created at far lower cost, helping to keep ahead of bacterial resistance.
To create new macrolides, the team reports, they used a method that allows for modular building, which they liken to the way cell phones are made—they start by building small-chemical blocks and then "weld" them together using a process that requires very few steps. They report also that the technique can be applied on a multigram scale, which means enough can be created at a time for use in experiments geared towards testing the results in killing bacteria.
To date, the team has produced 300 macrolides using their technique, which include some that have had already been created using the original method. They report that some have already been tested to see how well they fight bacteria and have met with some initial success. They acknowledge that a lot more work needs to be done to find out if drugs produced using the method are truly effective in fighting bacterial infections and if so, if they will prove to be safe for use in humans.
More information: Ian B. Seiple et al. A platform for the discovery of new macrolide antibiotics, Nature (2016). DOI: 10.1038/nature17967
Abstract
The chemical modification of structurally complex fermentation products, a process known as semisynthesis, has been an important tool in the discovery and manufacture of antibiotics for the treatment of various infectious diseases. However, many of the therapeutics obtained in this way are no longer effective, because bacterial resistance to these compounds has developed. Here we present a practical, fully synthetic route to macrolide antibiotics by the convergent assembly of simple chemical building blocks, enabling the synthesis of diverse structures not accessible by traditional semisynthetic approaches. More than 300 new macrolide antibiotic candidates, as well as the clinical candidate solithromycin, have been synthesized using our convergent approach. Evaluation of these compounds against a panel of pathogenic bacteria revealed that the majority of these structures had antibiotic activity, some efficacious against strains resistant to macrolides in current use. The chemistry we describe here provides a platform for the discovery of new macrolide antibiotics and may also serve as the basis for their manufacture.
Journal information: Nature
© 2016 Phys.org