Trimming piRNAs' tails to clip jumping genes' wings
A research group at the University of Tokyo has identified a Pac-Man-like enzyme called "Trimmer" involved in the generation of a class of small RNAs, which protect the genome of germ cells from unwanted genetic rewriting.
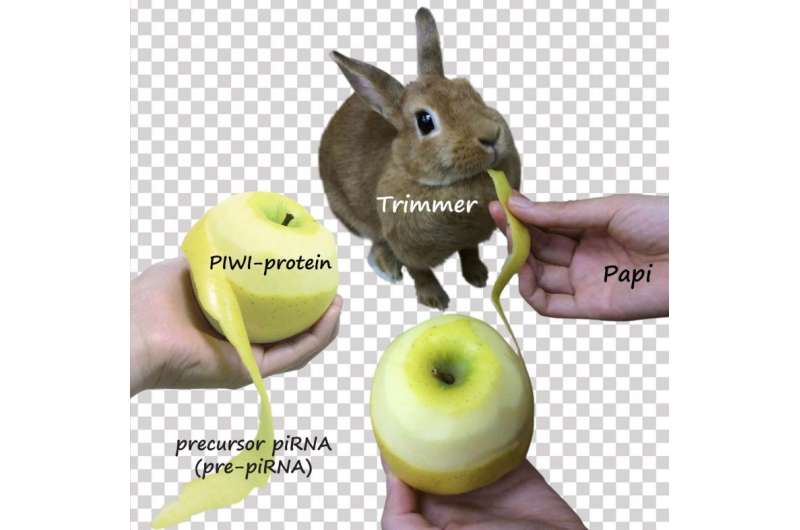
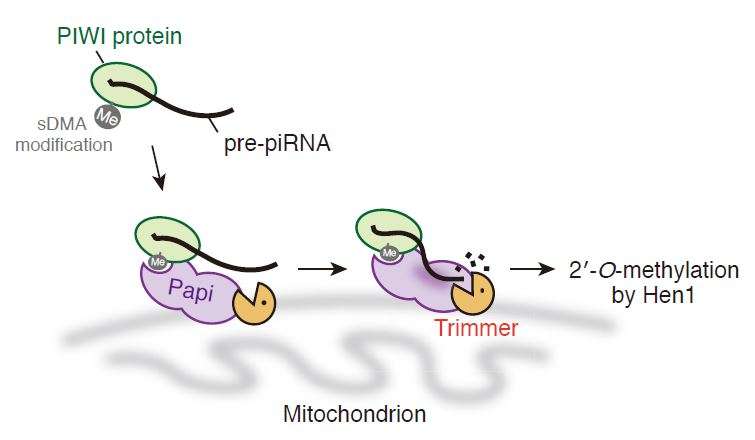
"Jumping genes" or transposons are small segments of DNA that can move around the genome. They can disrupt host genes and have been implicated in cancer and other diseases. As a result, organisms need to keep them under control, particularly in germ cells that give rise to sperm and eggs in animals to ensure the genome integrity of offspring. This is accomplished in germ cells by a class of small RNAs called piRNAs (PIWI-interacting RNAs), typically 24 to 30 nucleotides long, that suppress jumping gene expression. piRNAs are thought to be matured by trimming one end of their longer precursors (pre-piRNAs) to the final mature length. However, the enzyme responsible for this trimming process was unknown.
Research Associate Natsuko Izumi and Professor Yukihide Tomari at the University of Tokyo Institute for Molecular and Cellular Biosciences and their colleagues successfully identified a previously uncharacterized ribonuclease as the trimming protein "Trimmer" in silkworm ovary cells. Their data showed that Trimmer cannot act alone and requires Papi, a PIWI-associated protein, to trim the end of the pre-piRNAs. Moreover, they demonstrated that trimming of the pre-piRNAs is important for the function of piRNAs, and probably takes place on the surface of the mitochondria.
"We discovered the trimming activity in the pellet of silkworm cells back in 2011 and we knew the enzyme existed in the pellet. But, being insoluble, it was extremely difficult to identify Trimmer. In fact, we almost gave up many times", says Tomari. He continues, "A breakthrough came when we noticed the trimming activity is enriched in the mitochondria fraction of cells. We then patiently searched for conditions to solubilize the trimming activity from mitochondria. Even so, it still took another three years to identify Trimmer. The second breakthrough came when we realized that Trimmer partners with Papi. Discovering a nuclease that interacts with Papi and has pre-piRNA trimming activity, we were thrilled to find that it was the enzyme we had been searching for all along."
The study is published in the journal Cell.
More information: Cell, dx.doi.org/10.1016/j.cell.2016.01.008
Journal information: Cell
Provided by University of Tokyo