Graphics processors accelerate pattern discovery

Repeating patterns in complex biological networks can now be found hundreds of times faster using an algorithm that exploits the parallel computing capacity of modern graphics adapters. The A*STAR-led breakthrough opens the possibility of rapid genome scans for discrete molecular structures.
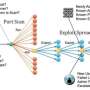
A network motif is a statistically significant connection of nodes that appears more frequently than chance would allow. In the gene transcription network for the bacteria Escherichia coli, for example, a simple network motif arises in the process by which a transcription factor responds to itself, known as negative auto-regulation. Such a motif occurs repeatedly in the transcriptome.
"This negative auto-regulation process has essential biological functions," explains Wenqing Lin from the A*STAR Institute for Infocomm Research. "Searching for these repeating 'graphs' is of particular importance in the study of network functionality."
These searches, however, are a significant computational drain and increasing the rate of motif discovery is a steep mathematical challenge. "Even state-of-the-art solutions require several days to derive network motifs from networks with only a few thousand vertices," says Lin.
The research team harnessed the capabilities of the modern computer graphics processors to overcome the limitations of existing network motif search algorithms. Graphics processing units often comprise thousands of computing cores, allowing Lin's team to adapt a large number of the computational tasks involved in graph mining. This can significantly reduce computation time.
By developing a parallelized code that utilizes the capabilities of graphical processing units (GPUs), including simultaneous execution of code on multiple GPU cores and efficient memory access patterns, the team was able to expedite the search by up to 100 times—the improved rate was confirmed through extensive experiments on a variety of biological networks.
The study demonstrates the feasibility of using GPUs for network motif search tasks. GPUs are also around 20 times cheaper than computer processors for equivalent performance, highlighting the potential for large-scale search projects.
Network motif discovery has many applications beyond biology, including pattern detection in digital circuits and other non-random networks. "Our research results can also be applied to solving the problems of enumerating all subgraphs of a large network, and finding the matches for a given subgraph in a large network," says Lin. "Both of these problems are fundamental aspects of graph mining and graph databases."
More information: "Network motif discovery: A GPU approach." IEEE 31st International Conference on Data Engineering (ICDE), 831–842 (2015). dx.doi.org/10.1109/ICDE.2015.7113337