First comprehensive analysis of the woolly mammoth genome completed

The first comprehensive analysis of the woolly mammoth genome reveals extensive genetic changes that allowed mammoths to adapt to life in the arctic. Mammoth genes that differed from their counterparts in elephants played roles in skin and hair development, fat metabolism, insulin signaling and numerous other traits. Genes linked to physical traits such as skull shape, small ears and short tails were also identified. As a test of function, a mammoth gene involved in temperature sensation was resurrected in the laboratory and its protein product characterized.
The study, published in Cell Reports on July 2, sheds light on the evolutionary biology of these extinct giants.
"This is by far the most comprehensive study to look at the genetic changes that make a woolly mammoth a woolly mammoth," said study author Vincent Lynch, PhD, assistant professor of human genetics at the University of Chicago. "They are an excellent model to understand how morphological evolution works, because mammoths are so closely related to living elephants, which have none of the traits they had."
Woolly mammoths last roamed the frigid tundra steppes of northern Asia, Europe and North America roughly 10,000 years ago. Well-studied due to the abundance of skeletons, frozen carcasses and depictions in prehistoric art, woolly mammoths possessed long, coarse fur, a thick layer of subcutaneous fat, small ears and tails and a brown-fat deposit behind the neck which may have functioned similar to a camel hump.
Previous efforts to sequence preserved mammoth DNA were error-prone or yielded insights into only a limited number of genes.
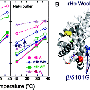
To thoroughly characterize mammoth-specific genes and their functions, Lynch and his colleagues deep sequenced the genomes of two woolly mammoths and three Asian elephants - the closest living relative of the mammoth. They then compared these genomes against each other and against the genome of African elephants, a slightly more distant evolutionary cousin to both mammoths and Asian elephants.
The team identified roughly 1.4 million genetic variants unique to woolly mammoths. These caused changes to the proteins produced by around 1,600 genes, including 26 that lost function and one that was duplicated. To infer the functional effects of these differences, they ran multiple computational analyses, including comparisons to massive databases of known gene functions and of mice in which genes are artificially deactivated.
Genes with mammoth-specific changes were most strongly linked to fat metabolism (including brown fat regulation), insulin signaling, skin and hair development (including genes associated with lighter hair color), temperature sensation and circadian clock biology - all of which would have been important for adapting to the extreme cold and dramatic seasonal variations in day length in the Arctic. The team also identified genes associated with the mammoth body plan, such as skull shape, small ears and short tails.
Of particular interest was the group of genes responsible for temperature sensation, which also play roles in hair growth and fat storage. The team used ancestral sequence reconstruction techniques to "resurrect" the mammoth version of one of these genes, TRPV3. When transplanted into human cells in the laboratory, the mammoth TRPV3 gene produced a protein that is less responsive to heat than an ancestral elephant version of the gene. This result is supported by observations in mice that have TRPV3 artificially silenced. These mice prefer colder environments than normal mice and have wavier hair.
Although the functions of these genes match well with the environment in which woolly mammoths were known to live, Lynch warns that it is not direct proof of their effects in live mammoths. The regulation of gene expression, for example, is extremely difficult to study through the genome alone.
"We can't know with absolute certainty the effects of these genes unless someone resurrects a complete woolly mammoth, but we can try to infer by doing experiments in the laboratory," he said. Lynch and his colleagues are now identifying candidates for other mammoth genes to functionally test as well as planning experiments to express mammoth proteins in elephant cells.
While his efforts are targeted toward understanding the molecular basis of evolution, Lynch acknowledges that the high-quality sequencing and analysis of woolly mammoth genomes can serve as a functional blueprint for efforts to "de-extinct" the mammoth.
"Eventually we'll be technically able to do it. But the question is: if you're technically able to do something, should you do it?" he said. "I personally think no. Mammoths are extinct and the environment in which they lived has changed. There are many animals on the edge of extinction that we should be helping instead."
More information: "Elephantid genomes reveal the molecular bases of Woolly Mammoth adaptations to the arctic," Cell Reports, 2015.
Journal information: Cell Reports
Provided by University of Chicago Medical Center