Horizontal gene transfer in E. coli
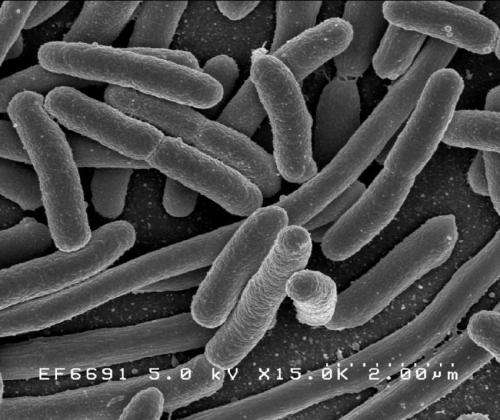
Escherichia coli O104 is an emergent disease-causing bacterium various strains of which are becoming increasingly well known and troublesome. The pathogen causes bloody diarrhea as well as and potentially fatal kidney damage, hemolytic uremic syndrome. Infection is usually through inadvertent ingestion of contaminated and incompletely cooked food or other materials, such as animals feces.
Escherichia coli is a so-called gram negative bacterium, commonly found in the intestine of humans and other mammals. Entero-hemorrhagic strains including O157, O26, O103 and O111 and specifically the sub-strain O157:H7 is an important cause of food borne illness in North America, the UK and Japan.
One particular strain, highlighted by Indian researchers in the International Journal of Bioinformatics Research and Applications, O104:H4, causes serious complications and has developed significant multiple-drug resistance to antibiotics. Moreover, it has acquired genes through horizontal transfer from other strains that make it even more virulent than others.
The team from Madurai Kamaraj University in Madurai, Tamil Nadu, working with colleagues at Genotypic Technology Pvt Ltd in Karnataka, have used the tools of computational molecular biology to identify 38 such horizontal gene transfer elements, prophage elements. These elements the team explains are genetic weapons that protect the bacteria from antibiotics and have been acquired from viruses, known as bacteriophages, that usually infect bacteria.
More than a quarter of the genome of this strain of E. coli comprises prophage elements, the team explains. These elements are also involved in the production of lethal compounds such as Shiga toxin, which give rise to many of the symptoms of infection. As such, they might represent new diagnostic markers or even targets for the development of novel antibiotics that circumvent the protective measures acquired by the bacteria.
More information: Kesavan, B., Srividhya, K.V., Krishnaswamy, S., Raja, M., Vidya, N. and Krishna Mohan, A.V.S.K. (2015) 'Understanding the virulence of the entero-aggregative E. coli O104:H4', Int. J. Bioinformatics Research and Applications, Vol. 11, No. 3, pp.187-199.
Provided by Inderscience Publishers