March 31, 2015 feature
Too many targets: Scientists create model to analyze ceRNA regulation, validate results with synthetic gene circuits
(Phys.org)—In the complex, somewhat rarified world of interactions between various flavors of RNA, one elusive goal is to understand the precise regulatory relationships between competing endogenous RNA (ceRNA), microRNA (miRNA), small interfering RNA (siRNA) and messenger RNA (mRNA).
To enter this world prepared, here's a quick overview:
- Competing endogenous RNAs, which include mRNAs, transcribed pseudogenes, long noncoding RNAs (lncRNA), and circular RNA (circRNA), regulate other RNA transcripts by competing for shared microRNA
- MicroRNA is a small non-coding RNA molecule containing about 22 nucleotides found in plants, animals, and some viruses, which functions in RNA silencing and post-transcriptional regulation of gene expression
- Messenger RNA is a large family of RNA molecules that convey genetic information from DNA to the ribosome, where they specify the amino acid sequence of the protein products of gene expression
- Pseudogenes are sections of a chromosome that are imperfect, dysfunctional copies of functional genes that have lost their protein-coding ability or are otherwise no longer expressed in the cell
- Long noncoding RNA (lncRNA) comprises a large and diverse class of transcribed RNA molecules with a length of more than 200 nucleotides that do not encode proteins, and whose expression is developmentally regulated and that can be tissue- and cell-type specific
- Circular RNA (circRNA) is a type of gene regulating noncoding RNA which, unlike the better-known linear RNA, forms a covalently closed continuous loop and that have not been shown to code for proteins
- Small interfering RNA (siRNA), sometimes known as short interfering RNA or silencing RNA, is a class of double-stranded RNA molecules, 20-25 base pairs in length, that has many functions but is most notable in the RNA interference (RNAi) pathway where it interferes with the expression of specific genes with complementary nucleotide sequences
Recently, scientists at Tsinghua University, Beijing investigated competing endogenous RNAs cross-regulation by creating a computational model, using it to quantitatively describe a minimum ceRNA network, and experimentally validating their model's predictions by utilizing multifluorescent synthetic gene circuits in cultured human cells. They found that the ceRNA effect is affected by the abundance of miRNA and ceRNAs, the number and affinity of binding sites, and the mRNA degradation pathway determined by the degree of miRNA/mRNA complementarity (a mirror image-like property shared between two DNA or RNA sequences that allows DNA replication and transcription). The researchers state that their findings have the potential to increase understanding quantitative properties of gene regulatory systems, contribute to the development of better tools for cell-type specific microRNA occupancy rate prediction, and benefit synthetic biology or genetic engineering research using miRNA or ceRNA in building more predictive parts with better quantitative behaviors.
Discussing the paper that he, Prof. Zhen Xie and their colleagues published in Proceedings of the National Academy of Sciences, Prof. Xiaowo Wang tells Phys.org that one of the main challenges the researchers faced was applying a model-guided synthetic biology approach to quantitatively analyze the behavior of miRNA-mediated ceRNA regulation. "In mammalian cells, each type of miRNA can interact with dozens to hundreds of target RNA species – including protein-coding mRNAs, long non-coding RNAs and recently-discovered circular RNAs." Moreover, he explains, each RNA species can also interact with multiple miRNA species through various miRNA regulatory elements, or MREs, and the complex interaction network of miRNAs and their target RNAs has been shown to allow indirect cross-regulation between different competing endogenous RNAs (ceRNAs) by sequestering shared miRNAs, which is essential for regulating many biological functions. "However," he points out, "natural microRNA-ceRNA networks are complex and hard to perturb, which makes it difficult to quantitatively understand the mechanism of miRNA-mediated ceRNA regulation. Thus, we decided to implement an artificial miRNA-ceRNA system by transfecting synthetic gene circuits into human HEK293 cells to simulate the behavior of ceRNA regulation." (Transfection is the process of deliberately introducing nucleic acids into cells.)
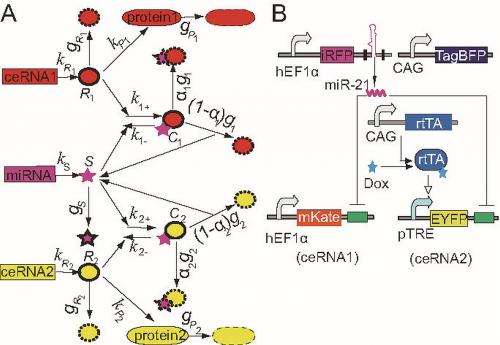
Wang adds that it was also necessary to establish a minimum ceRNA model to quantitatively analyze the behavior of ceRNA regulation. "Though several recent published mathematic models have revealed a titration mechanism underlying ceRNA regulation, many predictions deduced from the computational models have not been experimentally validated. In addition, it was also a significant challenge to validate the many parameters used in the mathematic model." To validate those predictions in turn required the implementation of multifluorescent synthetic gene circuits in cultured human cells.
"Thanks to the quick advance in genetic circuit engineering, people are now able to investigate complex gene regulations in a controlled and largely isolated biological setting. To mimic the minimal ceRNA model, we introduced a synthetic circuit that consists of five transcription units into mammalian cells by transient transfection followed by quantified fluorescent proteins by flow cytometry to analyze the circuit performance in individual cells." (In transient transfection, the nucleic acids introduced into the transfected cell are not permanently incorporated into the cellular genome, meaning that the effects of the nucleic acids within the cell last only a short amount of time.)
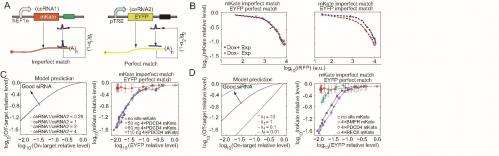
The scientists used three kinds of fluorescent proteins (iRFP, mKate and EYFP) to monitor the levels of miR-21, the first competing endogenous RNA (ceRNA1) and the second competing endogenous RNA (ceRNA2), respectively. "We also used another fluorescent protein, TagBFP, to gauge the variation in the transient transfection experiment," Wang adds. "In order to validate whether this experimental setup is suitable for quantitative measurements, we firstly confirmed that the iRFP, mKate, EYFP and TagBFP fluorescent intensities linearly correlated with the corresponding miRNA/mRNA levels. Then, we confirmed that both mKate and EYFP fluorescent intensities follow a linear relationship with TagBFP fluorescent intensity, suggesting that the TagBFP level is an appropriate indicator for the overall transcription rate in each transfection experiment. We were therefore confident that we were able to evaluate the underlying miRNA-ceRNA regulation under various experimental conditions by comparing the difference of the mKate level with or without induction of EYFP."
Relatedly, directly measuring the miRNA loss rate due to experimental difficulties was an issue. "The degree of complementarity of a miRNA/siRNA to its target site determines whether RISC complex enters into either miRNA-type or RNAi-type repression pathway," Wang tells Phys.org. (The RNA-Induced Silencing Complex is a multiprotein complex that incorporates one strand of a small siRNA or miRNA. RISC uses the siRNA or miRNA as a template for recognizing complementary mRNA.) "It's intriguing to study how these two types of regulations interact with each other when both regulations share the same miRNA/siRNA species." Wang points out that the miRNA loss rate α is thought to be different in these two types of regulations, which can strongly influence ceRNA crosstalk efficiency in the steady state – but the value of for miRNA regulation is largely unknown due to experimental difficulties in direct measurement.
In terms of the key insights, innovations and techniques that the researchers developed or leveraged to address these challenges, Wang says that in this study, the aim was to obtain a comprehensive and quantitative understanding of miRNA regulation principles on competing RNAs. "We first formulated a coarse-grained mathematical model for a minimum miRNA-ceRNA system composed of one miRNA species and two competing RNA targets. Then, we engineered and implemented a corresponding genetic circuit in cultured human embryonic kidney HEK293 cells to quantify the ceRNA effect under variable conditions by using a multi-fluorescent flow cytometry. Our results suggested that, by its competing nature, ceRNA network operates in a highly quantitative manner."
Through this process, the scientists were able to show that the miRNA level determines the regime where ceRNA effect occurs, and the elevation of miRNA level can shift the ceRNA-effective regime towards higher target transcriptional level. "In addition to an appropriate molecular environment, ceRNA-crosstalk efficiency also positively correlates with the binding strength of miRNA to its MREs, including the number of MREs and the thermodynamic binding stability, while keeping the effective regime unchanged relative to target transcriptional level," Wang continues. "We also found an nonreciprocal"– that is, one-way – "competing effect between partial and perfect complementary targets, which is largely due to the high miRNA recycling rate in RNAi-type regulation and the low miRNA recycling rate in miRNA-type regulation. In addition, we demonstrated that the RNA interference efficiency of small interference RNA (siRNA) can be significantly diminished by off-targets with high expression levels and strong binding sites." The researchers found that introducing an appropriate amount of the siRNA compensated for the effect caused by highly expressed off-targets, but not off-targets with strong binding sites.
The results presented in the paper suggest that the ceRNA effect is affected by several factors, including the abundance of microRNA (miRNA) and ceRNAs, the number and affinity of binding sites, and the mRNA degradation pathway determined by the degree of miRNA–mRNA complementarity. "Firstly, we found that the relative miRNA level determines the regime relative to ceRNA transcription level where the ceRNA effect occurs," Wang explains. "When the miRNA level is high, the cells need more target transcripts to trigger a ceRNA effect." In addition, he notes that they observed that ceRNA effect makes the target transcript more sensitive to microRNA repression when microRNA level is beyond a threshold.
"Secondly, when microRNA binds to their targets more tightly and when there are more microRNA binding sites in the target transcript, we observed a stronger ceRNA effect." However, he points out that unlike the relative miRNA level, the number and affinity of binding sites does not change the regime – whereas the ceRNA effect occurs.
"Lastly, MiRNA-mediated post-transcriptional regulation can be triggered by only 6-nt" (that is, six-nucleotide) "complementarity of the miRNA 5'-end so-called seed region to the target RNA. When miRNA perfectly matches the target RNA, the miRNA cleaves the target RNA and might be recycled to cleave the next target RNA; when miRNA imperfectly matches the target RNA, the miRNA binds to the target RNA, inhibiting translation or causing RNA destabilizing." To assay whether these two types of regulations interfere with each other when both regulations share the same miRNA/siRNA species, the scientists constructed a variant ceRNA system that consists of ceRNA1 with MREs partially paired to miR-21, and ceRNA2 with MREs perfectly paired to miR-21. (MREs, or microRNA response elements, are miRNA binding sites.)
"Interestingly," Wang reports, "ceRNA2 that contained partially paired MREs caused an obvious change on ceRNA1's miRNA dose–response curve, whereas introducing ceRNA2 with perfectly complementary MREs hardly affected miRNA repression efficacy on ceRNA1. This model simulation result suggested this nonreciprocal competition was mainly caused by the difference in the parameter α, which reflect the loss rate that a miRNA degraded together its target." By fitting their model to our experimental data, the researchers estimated that the loss rate for miRNA-type repression is about five times higher than that for RNAi-type repression in their experimental configuration.
The paper also sheds light on utilizing the scientists' competing model (based on rational design of effective siRNA) by finding that a nonreciprocal competing effect between partial and perfect complementary targets is mainly due to different miRNA loss rates in these two types of repressions. "We found that from a ceRNA effects perspective, a perfect pairing target – that is, an siRNA pathway – has little effect on imperfect paring target, while the existence of imperfect pairing target could influence the siRNA repression efficacy on perfect paring target. Such nonreciprocal effects are mainly due to the different loss rate of the two pathways according to our simulation analysis."
Finding these nonreciprocal effects inspired the scientists to conduct further analysis into the study-derived quantitative understanding of ceRNA cross-regulation via shared miRNA and implied an siRNA design strategy to reduce the siRNA off-target effect in mammalian cells. "According to our observations, the miRNA/siRNA balance has different repression efficiency for perfect and imperfect pairing targets at different miRNA/siRNA concentrations. For example," Wang illustrates, "at low miRNA concentrations, both perfect pairing and imperfect paring targets are both barely repressed – but at high miRNA concentrations, both kinds of targets are heavily repressed, while at mid-range concentrations repression efficacy is quite different again." Their goal is to use this property to find a proper siRNA concentration that can effectively repress perfect paring target, while keep the imperfect paring target largely unchanged.
In mapping an on-target/off-target relative repression curve to represent the relative repression efficiency on perfect paring target and imperfect paring target at different miRNA concentrations, the scientists found that the shape of the curve is influenced not by the relative expression level between on-target and off-target, but rather depends heavily on the relative binding efficiency. "In other words, in the case of high off-target expression level, we can increase the level of siRNA to compensate this effect to achieve the similar on-target-off-target repression ratio as the low off-target expression case. Moreover, the binding efficacy between siRNA and target off-target, which could be influenced by binding site number and hybridization energy, greatly influence the shape of the on-target/off-target relative repression curve. For off-target with high binding efficacy, the siRNA could hardly repress the on-target without touching the off-target. Thus we should avoid such strong off-target when we design siRNAs – but on the contrary, some weak off-target even with relatively high expression level could be tolerated."
A key outcome of the study was demonstrating how gene circuits offer a powerful tool, in a largely controlled manner, to understand the fundamental design principles of complex cellular systems. "Intrinsic gene networks are complex," Wang tells Phys.org. "They're often hard to perturb or manipulate. For example, the intrinsic miRNA-ceRNA network contains hundreds of types of miRNAs and thousands of types of target genes – and each miRNA could regulate hundreds of targets, and each target could bind by multiple types of miRNAs." In other words, it is almost impossible to perturb just a single ceRNA pair without touching the rest. However, he notes that synthetic circuits are relatively better isolated – and, importantly, parameters such as the expression level of miRNA or targets, number and strength of the binding sites can easily be manipulated, which is a great benefit in understanding quantitative properties of gene regulatory systems.
"According to our observations and another recent paper1, most mRNAs with moderate expression level could hardly induce significant gene expression changes through ceRNA effect. To that end, we're expanding and simulating our mathematical modeling to find if ceRNA effects could be more pronounced at certain conditions. In addition, were trying to incorporate these new observations into our previous published miRNA Occupancy Rate, or MIROR2, microRNA target prediction tool for better cell-type specific microRNA occupancy rate prediction."
The researchers are also considering using such systems and a synthetic biology approach to study the behavior of several signaling pathways. "We're interested in understanding the design principle of these regulatory systems and quantifying the information transformation through the pathways." As such, Wang notes that their study could benefit synthetic biology or genetic engineering research using miRNA or ceRNA as components for building more predictive parts with better quantitative behaviors.
More information: Model-guided quantitative analysis of microRNA-mediated regulation on competing endogenous RNAs using a synthetic gene circuit, Proceedings of the National Academy of Sciences (2015) 112(10):3158-3163, doi:10.1073/pnas.1413896112
Related:
1Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance, Molecular Cell (2014) 54(5):766–776, doi:10.1016/j.molcel.2014.03.045
2MIROR: a method for cell-type specific microRNA occupancy rate prediction, Molecular BioSystems (2014) 10:1377-1284, doi:10.1039/C3MB70610A
Journal information: Proceedings of the National Academy of Sciences , Molecular Cell
© 2015 Phys.org