Analyzing hundreds of cells in a few mouse-clicks
An EPFL PhD student has developed a technique for analyzing hundreds of images of cells in just a few mouse-clicks.
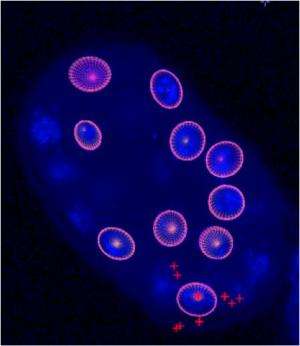
The increasingly powerful microscopes used in biomedical imaging provide biologists with 3D images of hundreds of cells, and cells in these images are often layered on each other. Under these conditions, it is impossible for traditional computational methods to determine the cells' properties (i.e., their size, shape, and density) quickly. Ricard Delgado-Gonzalo's work at EPFL is about to change that. The EPFL Biomedical Imaging Group (BIG) PhD student has developed a virtual tool that can analyze dozens of images in just an hour -which works out to hundreds of cells. All it takes is a standard computer.
The technique, called "Active Cell", is the fruit of complex mathematical calculations and sophisticated algorithms. Seen on a computer display it looks like a flattened sphere that can be squished into various shapes; it has lines around its surface, rather like a world-map globe. Users can scroll their mouse over a 3D digital microscopic image, using the globe to detect and segregate single cells from within the image -even if the cells are in an environment with many other surrounding objects. The globe precisely anchors itself to the target cell and adopts its contours, determining the cell's shape, size and density in real time. This can be extremely useful in situations where cells are undergoing therapeutic treatments and require monitoring, to take just one example. The technique, which will soon be written up in IEEE Transactions on Image Processing, has multiple applications: for instance, it could also be used to analyze imaging of organs, or any other body that has a more or less ellipsoid shape.
Stable, extremely precise data
"Until now, biologists have had to manually surround the cell they wanted to analyze by clicking over and over again on its perimeter points, kind of like you do in Photoshop," says Delgado-Gonzalo. "With this method, all you need to do is drag the globe near the target cell, click on it, and it's automatically analyzed."
Available as freeware
In order to make their results as widely available as possible, the EPFL team has put their solution on the open-source platform Icy. This platform contains the equivalent of an "app store" for biomedical imaging. And in fact biologists were consulted as the tool was being developed. A 2D version is already being used by scientists, while the team perfects an upcoming 3D version. "In addition, we're currently designing a website user-guide and a tablets version of the tool," says Delgado-Gonzalo.
Strong reviews from the scientific community
The new "snake," whose margin of error is on the order of one tenth of a pixel, has been very favorably received by the scientific community. "I used it for my Master's project when I was at the EPFL," says Mustapha Al Kharfane, now an engineer at Biosensors Europe SA. "We analyzed an MRI of a patient's aorta to determine its elasticity and blood-flow rates. Those are two key clinical parameters for assessing a person's cardiovascular risk. The 'snake' saved me hours of work."
Another user, Nicolas Dénervaud, is working on a PhD at EPFL's Laboratory of Biological Network Characterization (LBNC). He is equally positive, commenting, "We use the new technique to analyze large numbers of yeast cells in maximally dense samples. The algorithm is fast and robust."
More information: bigwww.epfl.ch/preprints/delgadogonzalo1302p.pdf
Provided by Ecole Polytechnique Federale de Lausanne