Technique quickly identifies bacteria for food safety
Researchers at Purdue University have used a new technique to rapidly detect and precisely identify bacteria, including dangerous E. coli, without time-consuming treatments usually required.
The technique, called desorption electrospray ionization, or DESI, could be used to create a new class of fast, accurate detectors for applications ranging from food safety to homeland security, said R. Graham Cooks, the Henry Bohn Hass Distinguished Professor of Chemistry in Purdue's College of Science.
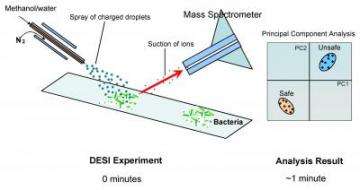
Using a mass spectrometer to analyze bacteria and other microorganisms ordinarily takes several hours and requires that samples be specially treated and prepared in a lengthy series of steps. DESI eliminates the pretreatment steps, enabling researchers to take "fingerprints" of bacteria in less than a minute using a mass spectrometer.
"This is the first time we've been able to chemically analyze and accurately identify the type of bacteria using a mass spectrometer without any prior pretreatment within a matter of seconds," Cooks said.
New findings show how the Purdue researchers used the method to detect living, untreated bacteria, including E. coli and Salmonella typhimurium, both of which cause potentially fatal infections in humans.
"There is always an advantage to the analysis of living systems because the bacteria retain their original properties," Cooks said.
The findings are detailed in a paper appearing Jan. 7 in the journal Chemical Communications. The paper was written by chemistry graduate students Yishu Song, Nari Talaty and Zhengzheng Pan; Andy W. Tao, an assistant professor of biochemistry; and Cooks.
Mass spectrometry works by turning molecules into ions, or electrically charged versions of themselves, inside the instrument's vacuum chamber. Once ionized, the molecules can be more easily manipulated, detected and analyzed based on their masses. The key DESI innovation is performing the ionization step in the air or directly on surfaces outside of the mass spectrometer's vacuum chamber. When combined with portable mass spectrometers also under development at Purdue, DESI promises to provide a new class of compact detectors.
Purdue researchers are focusing on three potential applications for detecting and identifying pathogens: food safety, medical analysis and homeland security. Such a detector could quickly analyze foods, medical cultures and the air in hospitals, subway stations and airports, Cooks said.
The researchers are able to detect one nanogram, or a billionth of a gram, of a particular bacterium. More importantly, the method enables researchers to identify a particular bacterium down to its subspecies, a level of accuracy needed to detect and track infectious pathogens. The identifications are based on specific chemical compounds, called lipids and fatty acids, in the bacteria.
"We can determine the subspecies and glean other information by looking at the pattern of chemicals making up the pathogen, a sort of fingerprint revealed by mass spectrometry," Cooks said. "Conventional wisdom says quick methods such as ours will not be highly chemically or biologically specific, but we have proven that this technique is extremely accurate."
The procedure involves spraying water in the presence of an electric field, causing water molecules to become positively charged "hydronium ions," which contain an extra proton. When the positively charged droplets come into contact with the sample being tested, the hydronium ions transfer their extra proton to molecules in the sample, turning them into ions. The ionized molecules are then vacuumed from the surface into the mass spectrometer, where the masses of the ions are measured and the material analyzed.
Such a system could alert employees in the food and health-care industries to the presence of pathogens and could provide security personnel with a new tool for screening suspicious suitcases or packages.
Song will further the research, conducting experiments to look for bacterial contaminants in foods. Ongoing work by Talaty with international E. coli expert Barry Wanner, a professor in Purdue's Department of Biological Sciences, will apply the method to living bacteria in so-called biofilms.
DESI has been commercialized by Indianapolis-based Prosolia Inc.
"This method could be applied very soon because the hardware is already available," Cooks said.
The DESI instrument and mass spectrometer used in the research are housed at the Bindley Bioscience Center at Purdue's Discovery Park.
Cooks also is leading work to build miniature versions of the normally bulky mass spectrometer, creating shoebox-size instruments that weigh about 10 kilograms (22 pounds), compared to about 30 times that weight for a conventional mass spectrometer.
Source: Purdue University