Histone modifications control accessibility of DNA
(PhysOrg.com) -- n an advanced online publication in Nature Structural & Molecular Biology scientist from Dirk Schübeler's group from the Friedrich Miescher Institute for Biomedical Research provide a genome-wide view of histone modifications and its influence on DNA accessibility. Their results expand the established break-up of chromatin into euchromatin and heterochomatin to a more complex pattern and link it to histone modifications.
DNA needs to be accessible. Otherwise DNA-based reactions like transcription, replication and repair will not happen. But DNA also needs to fit into the tiny cell nucleus, so a certain amount of compaction is necessary. The cells achieve this compaction through histones, which act as spools around which the DNA winds, sometimes tighter, other times looser. For a while now, scientists have hypothesized that regulatory mechanisms involving histones control accessibility to DNA. Adding varying numbers of methyl and acetyl residues to histones, was thought to regulate not only the higher order structure of the genetic material but also recruitment of proteins to the DNA. But do they also modulate DNA accessibility?
Dirk Schübeler and his research group at the Friedrich Miescher Institute for Biomedical Research were now able to show for the first time a clear link between DNA accessibility and histone modifications in the fruit fly Drosophila melanogaster.
In an advanced online publication in Nature Structural & Molecular Biology they provide a genome-wide view of histone modifications and its influence on DNA accessibility. Using a modified DNA methylase accessibility assay, they could distinguish between different levels of chromatin structures associated with distinct histone modification patterns.
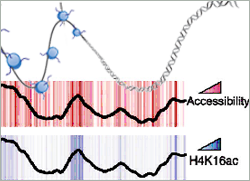
Accessible DNA is rich in methylated histone H3 (H3K4me) and hyperacetylated histone H4 (H4K16ac). These histones localize to gene rich areas, active promoters and origins of replication where the DNA is highly exposed. On the other hand, DNA is hardly accessible whenever histone 3 is methylated on lysine residue 27. These H3K27me3-rich areas cluster to large, inactive chromosomal regions. Since the Polycomb family of proteins is known to repress transcription and to set methylation on H3K27 it is conceivable that in H3K27me3 areas the observed reduced accessibility is mediating gene silencing.
"We show that there are indeed differences in DNA accessibility over large chromosomal regions, reflecting most likely various degrees of chromatin compaction. These differences correlate with distinct histone modification patterns," comments Dirk Schübeler. "This challenges the simple breakup of chromatin into active euchromatin and inactive heterochromatin. Our findings reveal a more complex pattern of genome accessibility linked to histone modifications."
More information: Publication in Nature Structural & Molecular Biology - www.nature.com/nsmb/journal/va … t/abs/nsmb.1825.html
Provided by Friedrich Miescher Institute for Biomedical Research














