Sulphur-eating bacteria limit acid run-off and CO2
(PhysOrg.com) -- Acid Mine Drainage (AMD) is caused when sulphur in mine tailings reacts with water and oxygen in the environment to produce sulphuric acid. It is a major environmental issue, with AMD a concern for lake acidification and water quality. AMD is also implicated as a climate change culprit - the sulphuric acid dissolves carbonate minerals in the underlying rock, liberating carbon dioxide in a process known as acid rock weathering.
Using two beamlines at the Canadian Light Source (CLS) and a third at the Advanced Light Source (ALS), researchers from McMaster University have found that two species of bacteria isolated from a mine tailings pond in northern Ontario actually work together to limit the amount of acid produced by sharing the sulphur in the tailings as an energy source.
The discovery, made by Ph.D. student Kelsey Norlund and her advisor Lesley Warren with the help of veteran synchrotron researcher Adam Hitchcock, not only demonstrates how bacteria can modify their environment, but could also lead to a rethink of the amount of atmospheric carbon produced by acid weathering that is included in global models by as much as 26 million tonnes.
“Before this work, our understanding of how this system operates was based largely on chemical models of water quality in these tailings ponds; things like the sulphur chemistry and pH,” explains Warren. “Everyone knew that bacteria were there and playing a role, but that role was not well described beyond broad brushstrokes.”
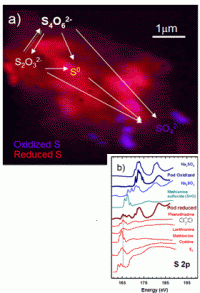
The bacteria, strains of Acidithiobacillus ferrooxidans and Acidiphilium spp., work together to form microscopic pods, with A. ferrooxidans on the outside and Acidiphilium on the inside. While both microbes can use elemental sulphur as an energy source, Norlund and Warren found that only A. ferrooxidans makes use of the raw sulphur, producing different chemical forms that were then metabolized by the Acidiphilium cells, producing sulphur compounds that, in turn, could be reused by
A. ferrooxidans. The result: a microscopic geochemical reactor, in which each species of microbe feeds off the sulphur-containing by-products of the other, rather than competing for the same chemical food source.
“We were able to see how these different bacteria organize the different types of sulphur,” says Norlund. “It’s a highly organized, cooperative ecological partnership. The waste products of one strain are food for the other, but they aren’t just using the leftovers - they’re actually regenerating each other’s food source.”
The cycling of sulphur compounds in this manner markedly reduces the amount of sulphur available to form sulphuric acid, dissolve carbonate rocks and release CO2. Norlund estimates that between 40 and 90 percent less carbon is released into the atmosphere because of this cycling than has been predicted based on the sulphur chemistry alone.
The next steps for the research team include more beam time at the CLS to study how the microbial pods form in the first place, as well as going out into the wider environment to identify other such partnerships and assess the roles they play in global geochemical cycles.
“We suspect that this sort of relationship is quite prevalent,” says Warren. “It’s just a matter of knowing where to look and what to look for.”
More information: Norlund et al. 2009. Microbial Architecture of Environmental Sulfur Processes: A Novel Syntrophic Sulfur-Metabolizing Consortia. Environmental Science and Technology 43, pp. 8781-8786. DOI:10.1021/es803616k
Provided by Canadian Light Source















