Modeling Genomic Erosion
(PhysOrg.com) -- Even though scientists have successfully sequenced the human genome, they still lack a clear picture of exactly how coding and non-coding DNA sequences function together, or how genomes evolve over time. This has been particularly problematic for scientists who use genetic similarities to characterize evolutionary divergence. Historically, the processes of genetic evolution and genome degradation have been difficult to study due to technological limitations and lack of accurate historical records for species.
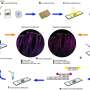
Researchers are now beginning to understand this process using symbiotic bacteria as model systems. When these bacteria went from free-living to obligate symbionts (unable to survive apart from the host), there were a number of genes that the bacteria no longer needed because they could obtain the necessary cofactors from the host species and because fewer genes are maintained in small populations such as those of symbionts. These genes were more susceptible to mutation and genetic erosion that ultimately resulted in genome reduction because they were no longer necessary for the bacteria to survive.
A team lead by Nancy Moran of the University of Arizona recently published data that characterize the dynamics and timescale of genomic erosion of the symbiotic bacteria of pea aphids. Pea aphids, native to Eurasia, were introduced to North America in the 1870s.
"We have good historical records for this species," said Moran. "Because this particular aphid species was not native to North America we know that the upper time limit for genetic divergence of the symbiotic bacteria descending from a single aphid introduction is 135 years or less."
The researchers sequenced the entire genome of seven different strains of symbiotic bacteria found within pea aphid strains in North America, including two strains that diverged in the laboratory. Extensive statistical analysis revealed mutation rates 10 times higher than the highest estimate for any species of bacteria. The authors used these results to construct a model of the stepwise process that eventually leads to genome reduction. This work is significant as the basis for how the process of genomic erosion occurs and provides insights about how this mechanism may act on a larger scale.
A full account of this project is published in the January 16 issue of Science magazine.
"Previous National Science Foundation-funded studies examined mutation rates and processes over very long periods of evolutionary time," stated Moran. "This work contributes to our understanding of whole-genome mutation rates and patterns over a well-defined, short period of time, allowing more exact reconstruction of changes. Our model is also consistent with other evidence characterizing mutational patterns."
Provided by National Science Foundation