MIT creates 3-D images of living cell
A new imaging technique developed at MIT has allowed scientists to create the first 3D images of a living cell, using a method similar to the X-ray CT scans doctors use to see inside the body.
The technique, described in a paper published in the Aug. 12 online edition of Nature Methods, could be used to produce the most detailed images yet of what goes on inside a living cell without the help of fluorescent markers or other externally added contrast agents, said Michael Feld, director of MIT's George R. Harrison Spectroscopy Laboratory and a professor of physics.
“Accomplishing this has been my dream, and a goal of our laboratory, for several years,” said Feld, senior author of the paper. “For the first time the functional activities of living cells can be studied in their native state.”
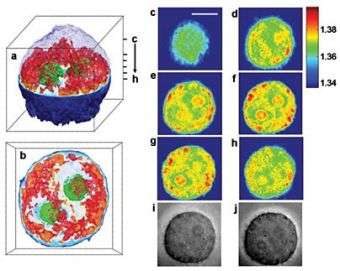
Using the new technique, his team has created three-dimensional images of cervical cancer cells, showing internal cell structures. They've also imaged C. elegans, a small worm, as well as several other cell types.
The researchers based their technique on the same concept used to create three-dimensional CT (computed tomography) images of the human body, which allow doctors to diagnose and treat medical conditions. CT images are generated by combining a series of two-dimensional X-ray images taken as the X-ray source rotates around the object.
“You can reconstruct a 3D representation of an object from multiple images taken from multiple directions,” said Wonshik Choi, lead author of the paper and a Spectroscopy Laboratory postdoctoral associate.
Cells don't absorb much visible light, so the researchers instead created their images by taking advantage of a property known as refractive index. Every material has a well-defined refractive index, which is a measure of how much the speed of light is reduced as it passes through the material. The higher the index, the slower the light travels.
The researchers made their measurements using a technique known as interferometry, in which a light wave passing through a cell is compared with a reference wave that doesn't pass through it. A 2D image containing information about refractive index is thus obtained.
To create a 3D image, the researchers combined 100 two-dimensional images taken from different angles. The resulting images are essentially 3D maps of the refractive index of the cell's organelles. The entire process took about 10 seconds, but the researchers recently reduced this time to 0.1 seconds.
The team's image of a cervical cancer cell reveals the cell nucleus, the nucleolus and a number of smaller organelles in the cytoplasm. The researchers are currently in the process of better characterizing these organelles by combining the technique with fluorescence microscopy and other techniques.
“One key advantage of the new technique is that it can be used to study live cells without any preparation,” said Kamran Badizadegan, principal research scientist in the Spectroscopy Laboratory and assistant professor of pathology at Harvard Medical School, and one of the authors of the paper. With essentially all other 3D imaging techniques, the samples must be fixed with chemicals, frozen, stained with dyes, metallized or otherwise processed to provide detailed structural information.
“When you fix the cells, you can't look at their movements, and when you add external contrast agents you can never be sure that you haven't somehow interfered with normal cellular function,” said Badizadegan.
The current resolution of the new technique is about 500 nanometers, or billionths of a meter, but the team is working on improving the resolution. “We are confident that we can attain 150 nanometers, and perhaps higher resolution is possible,” Feld said. “We expect this new technique to serve as a complement to electron microscopy, which has a resolution of approximately 10 nanometers.”
Source: Massachusetts Institute of Technology




















